The nucleolus (/njuːˈkliːələs, ˌnjuːkliˈoʊləs/; pl.: nucleoli /-laɪ/) is the largest structure in the nucleus of eukaryotic cells.[1] It is best known as the site of ribosome biogenesis, which is the synthesis of ribosomes. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress.[2] Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli can be the cause of several human conditions called "nucleolopathies"[3][4] and the nucleolus is being investigated as a target for cancer chemotherapy.[5][6]
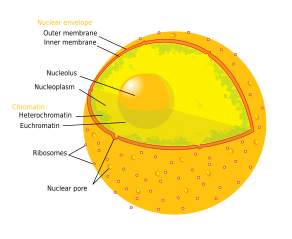
| Cell biology | |
|---|---|
| Animal cell diagram | |
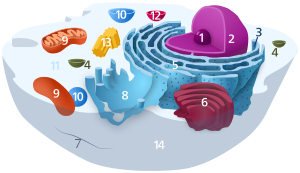 Components of a typical animal cell:
|
History
The nucleolus was identified by bright-field microscopy during the 1830s.[7] Theodor Schwann in his 1939 treatise describes that Schleiden had identified small corpuscleus in nuclei, and names the structure "Kernkörperchen". In a 1947 translation of the work to English, the structure receives the name of "nucleolus".[8][9]
In addition to these peculiarities of the cytoblast, already made known by Brown and Meyen, Schleiden has discovered in its interior a small corpuscle (see plate I, fig. 1, 4,) which, in the fully-developed cytoblast, looks like a thick ring, or a thick-walled hollow globule. It appears, however, to present a different appearance in different cytoblasts. Sometimes only the external sharply-defined circle of this ring can be distinguished, with a dark point in the centre,—occasionally, and indeed most frequently, only a sharply circumscribed spot. In other instances this spot is very small, and sometimes cannot be recognized at all. As it will frequently be necessary to speak of this body in the following treatise, I will for brevity’s sake name it the “nucleolus,” (Kernkorperchen, ‘nucleus-corpuscle.”)
— Theodor Schwann, translated by Henry Smith, Microscopical Researches Into the Accordance in the Structure and Growth of Animals and Plants, page 3
Little was known about the function of the nucleolus until 1964, when a study[10] of nucleoli by John Gurdon and Donald Brown in the African clawed frog Xenopus laevis generated increasing interest in the function and detailed structure of the nucleolus. They found that 25% of the frog eggs had no nucleolus, and that such eggs were not capable of life. Half of the eggs had one nucleolus and 25% had two. They concluded that the nucleolus had a function necessary for life. In 1966, Max L. Birnstiel and collaborators showed via nucleic acid hybridization experiments that DNA within nucleoli codes for ribosomal RNA.[11][12]
Structure
Three major components of the nucleolus are recognized: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC).[1] Transcription of the rDNA occurs in the FC.[13] The DFC contains the protein fibrillarin,[13] which is important in rRNA processing. The GC contains the protein nucleophosmin,[13] (B23 in the external image), which is also involved in ribosome biogenesis.
However, it has been proposed that this particular organization is only observed in higher eukaryotes and that it evolved from a bipartite organization with the transition from anamniotes to amniotes. Reflecting the substantial increase in the DNA intergenic region, an original fibrillar component would have separated into the FC and the DFC.[14]
Another structure identified within many nucleoli (particularly in plants) is a clear area in the center of the structure referred to as a nucleolar vacuole.[15]Nucleoli of various plant species have been shown to have very high concentrations of iron[16] in contrast to human and animal cell nucleoli.
The nucleolus ultrastructure can be seen through an electron microscope, while the organization and dynamics can be studied through fluorescent protein tagging and fluorescent recovery after photobleaching (FRAP). Antibodies against the PAF49 protein can also be used as a marker for the nucleolus in immunofluorescence experiments.[17]
Although usually only one or two nucleoli can be seen, a diploid human cell has ten nucleolus organizer regions (NORs) and could have more nucleoli. Most often multiple NORs participate in each nucleolus.[18]
Function and ribosome assembly

In ribosome biogenesis, two of the three eukaryotic RNA polymerases (Pol I and Pol III) are required, and these function in a coordinated manner. In an initial stage, the rRNA genes are transcribed as a single unit within the nucleolus by RNA polymerase I. In order for this transcription to occur, several pol I-associated factors and DNA-specific trans-acting factors are required. In yeast, the most important are: UAF (upstream activating factor), TBP (TATA-box binding protein), and core binding factor (CBF), which bind promoter elements and form the preinitiation complex (PIC), which is in turn recognized by RNA polymerase. In humans, a similar PIC is assembled with SL1, the promoter selectivity factor (composed of TBP and TBP-associated factors, or TAFs), transcription initiation factors, and UBF (upstream binding factor). RNA polymerase I transcribes most rRNA transcripts (28S, 18S, and 5.8S), but the 5S rRNA subunit (component of the 60S ribosomal subunit) is transcribed by RNA polymerase III.[19]
Transcription of rRNA yields a long precursor molecule (45S pre-rRNA), which still contains the ITS and ETS. Further processing is needed to generate the 18S RNA, 5.8S, and 28S RNA molecules. In eukaryotes, the RNA-modifying enzymes are brought to their respective recognition sites by interaction with guide RNAs, which bind these specific sequences. These guide RNAs belong to the class of small nucleolar RNAs (snoRNAs), which are complexed with proteins and exist as small-nucleolar-ribonucleoproteins (snoRNPs). Once the rRNA subunits are processed, they are ready to be assembled into larger ribosomal subunits. However, an additional rRNA molecule, the 5S rRNA, is also necessary. In yeast, the 5S rDNA sequence is localized in the intergenic spacer and is transcribed in the nucleolus by RNA polymerase.
In higher eukaryotes and plants, the situation is more complex, for the 5S DNA sequence lies outside the NOR and is transcribed by RNA Pol III in the nucleoplasm, after which it finds its way into the nucleolus to participate in the ribosome assembly. This assembly not only involves the rRNA, but also ribosomal proteins. The genes encoding these r-proteins are transcribed by Pol II in the nucleoplasm by a "conventional" pathway of protein synthesis (transcription, pre-mRNA processing, nuclear export of mature mRNA, and translation on cytoplasmic ribosomes). The mature r-proteins are then imported into the nucleus and, finally, the nucleolus. Association and maturation of rRNA and r-proteins result in the formation of the 40S (small) and 60S (large) subunits of the complete ribosome. These are exported through the nuclear pore complexes to the cytoplasm, where they remain free or become associated with the endoplasmic reticulum, forming the rough endoplasmic reticulum (RER).[20][21]
In human endometrial cells, a network of nucleolar channels is sometimes formed. The origin and function of this network have not yet been clearly identified.[22]
Sequestration of proteins
In addition to its role in ribosomal biogenesis, the nucleolus is known to capture and immobilize proteins, a process known as nucleolar detention. Proteins that are detained in the nucleolus are unable to diffuse and to interact with their binding partners. Targets of this post-translational regulatory mechanism include VHL, PML, MDM2, POLD1, RelA, HAND1 and hTERT, among many others. It is now known that long noncoding RNAs originating from intergenic regions of the nucleolus are responsible for this phenomenon.[23]
See also
References
Further reading
- Cooper GM (2000). "The Nucleolus". The Cell: A Molecular Approach (2nd ed.). Sunderland MA: Sinauer Associates. ISBN 978-0-87893-106-4.
- Tiku V, Antebi A (August 2018). "Nucleolar Function in Lifespan Regulation". Trends in Cell Biology. 28 (8): 662–672. doi:10.1016/j.tcb.2018.03.007. PMID 29779866. S2CID 29167518.
- JoAnna Klein (20 May 2018). "The Thing Inside Your Cells That Might Determine How Long You Live". The New York Times.
External links

- Nucleolus under electron microscope II at uni-mainz.de
- Nuclear Protein Database – search under compartment
- Cell+Nucleolus at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Histology image: 20104loa – Histology Learning System at Boston University