Rotaviruses are the most common cause of diarrhoeal disease among infants and young children.[1] Nearly every child in the world is infected with a rotavirus at least once by the age of five.[2] Immunity develops with each infection, so subsequent infections are less severe. Adults are rarely affected.[3] Rotavirus is a genus of double-stranded RNA viruses in the family Reoviridae. There are nine species of the genus, referred to as A, B, C, D, F, G, H, I and J. Rotavirus A is the most common species, and these rotaviruses cause more than 90% of rotavirus infections in humans.[4]
| Rotavirus | |
|---|---|
 | |
| Computer–aided reconstruction of a rotavirus based on several electron micrographs | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Duplornaviricota |
| Class: | Resentoviricetes |
| Order: | Reovirales |
| Family: | Sedoreoviridae |
| Subfamily: | Sedoreovirinae |
| Genus: | Rotavirus |
| Species | |
| |
The virus is transmitted by the faecal–oral route. It infects and damages the cells that line the small intestine and causes gastroenteritis (which is often called "stomach flu" despite having no relation to influenza). Although rotavirus was discovered in 1973 by Ruth Bishop and her colleagues by electron micrograph images[5] and accounts for approximately one third of hospitalisations for severe diarrhoea in infants and children,[6] its importance has historically been underestimated within the public health community, particularly in developing countries.[7] In addition to its impact on human health, rotavirus also infects other animals, and is a pathogen of livestock.[8]
Rotaviral enteritis is usually an easily managed disease of childhood, but among children under 5 years of age rotavirus caused an estimated 151,714 deaths from diarrhoea in 2019.[9] In the United States, before initiation of the rotavirus vaccination programme in the 2000s, rotavirus caused about 2.7 million cases of severe gastroenteritis in children, almost 60,000 hospitalisations, and around 37 deaths each year.[10] Following rotavirus vaccine introduction in the United States, hospitalisation rates have fallen significantly.[11][12] Public health campaigns to combat rotavirus focus on providing oral rehydration therapy for infected children and vaccination to prevent the disease.[13] The incidence and severity of rotavirus infections has declined significantly in countries that have added rotavirus vaccine to their routine childhood immunisation policies.[14][15][16]
Virology
Types of rotavirus
There are nine species of rotavirus (sometimes informally called groups) referred to as A, B, C, D, F, G, H, I and J.[17][18] Humans are primarily infected by rotaviruses in the species Rotavirus A. A–I species cause disease in other animals,[19] species H in pigs, D, F and G in birds, I in cats and J in bats.[20][21][22][23]
Within group A rotaviruses there are different strains, called serotypes.[24] As with influenza virus, a dual classification system is used based on two proteins on the surface of the virus. The glycoprotein VP7 defines the G serotypes and the protease-sensitive protein VP4 defines P serotypes.[25] Because the two genes that determine G-types and P-types can be passed on separately to progeny viruses, different combinations are found.[25] A whole genome genotyping system has been established for group A rotaviruses, which has been used to determine the origin of atypical strains.[26] The prevalence of the individual G-types and P-types varies between, and within, countries and years.[27] There are at least 36 G types and 51 P types[28] but in infections of humans only a few combinations of G and P types predominate. They are G1P[8], G2P[4], G3P[8], G4P[8], G9P[8] and G12P[8].[29]
Structure
The genome of rotaviruses consists of 11 unique double helix molecules of RNA (dsRNA) which are 18,555 nucleotides in total. Each helix, or segment, is a gene, numbered 1 to 11 by decreasing size. Each gene codes for one protein, except genes 9, which codes for two.[30] The RNA is surrounded by a three-layered icosahedral protein capsid. Viral particles are up to 76.5 nm in diameter[31][32] and are not enveloped.[33]
Proteins
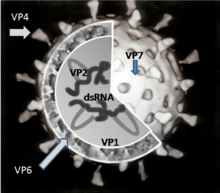
There are six viral proteins (VPs) that form the virus particle (virion). These structural proteins are called VP1, VP2, VP3, VP4, VP6 and VP7. In addition to the VPs, there are six nonstructural proteins (NSPs), that are only produced in cells infected by rotavirus. These are called NSP1, NSP2, NSP3, NSP4, NSP5 and NSP6.[19]
At least six of the twelve proteins encoded by the rotavirus genome bind RNA.[35] The role of these proteins in rotavirus replication is not entirely understood; their functions are thought to be related to RNA synthesis and packaging in the virion, mRNA transport to the site of genome replication, and mRNA translation and regulation of gene expression.[36]
Structural proteins
VP1 is located in the core of the virus particle and is an RNA-dependent RNA polymerase enzyme.[37] In an infected cell this enzyme produces mRNA transcripts for the synthesis of viral proteins and produces copies of the rotavirus genome RNA segments for newly produced virus particles.[38]
VP2 forms the core layer of the virion and binds the RNA genome.[39]
VP3 is part of the inner core of the virion and is an enzyme called guanylyl transferase. This is a capping enzyme that catalyses the formation of the 5' cap in the post-transcriptional modification of mRNA.[40] The cap stabilises viral mRNA by protecting it from nucleic acid degrading enzymes called nucleases.[41]
VP4 is on the surface of the virion that protrudes as a spike.[42] It binds to molecules on the surface of cells called receptors and drives the entry of the virus into the cell.[43] VP4 has to be modified by the protease enzyme trypsin, which is found in the gut, into VP5* and VP8* before the virus is infectious.[44] VP4 determines how virulent the virus is and it determines the P-type of the virus.[45] In humans there is an association between the blood group (Lewis antigen system, ABO blood group system and secretor status) and susceptibility to infection. Non-secretors seem resistant to infection by types P[4] and P[8], indicating that blood group antigens are the receptors for these genotypes.[46] This resistance is dependent on the rotavirus genotype.[47]
VP6 forms the bulk of the capsid. It is highly antigenic and can be used to identify rotavirus species.[48] This protein is used in laboratory tests for rotavirus infections.[49]
VP7 is a glycoprotein that forms the outer surface of the virion. Apart from its structural functions, it determines the G-type of the strain and, along with VP4, is involved in immunity to infection.[31]
Nonstructural viral proteins
NSP1, the product of gene 5, is a nonstructural RNA-binding protein.[50] NSP1 also blocks the interferon response, the part of the innate immune system that protects cells from viral infection. NSP1 causes the proteosome to degrade key signaling components required to stimulate production of interferon in an infected cell and to respond to interferon secreted by adjacent cells.
Targets for degradation include several IRF transcription factors required for interferon gene transcription.[51]
NSP2 is an RNA-binding protein that accumulates in cytoplasmic inclusions (viroplasms) and is required for genome replication.[52][39]
NSP3 is bound to viral mRNAs in infected cells and it is responsible for the shutdown of cellular protein synthesis.[53] NSP3 inactivates two translation initiation factors essential for synthesis of proteins from host mRNA.
First, NSP3 ejects poly(A)-binding protein (PABP) from the translation initiation factor eIF4F. PABP is required for efficient translation of transcripts with a 3' poly(A) tail, which is found on most host cell transcripts. Second, NSP3 inactivates eIF2 by stimulating its phosphorylation.[54] Efficient translation of rotavirus mRNA, which lacks the 3' poly(A) tail, does not require either of these factors.[55]
NSP4 is a viral enterotoxin that induces diarrhoea and was the first viral enterotoxin discovered.[56] It is a viroporin that elevates cytosolic Ca2+ in mammalian cells.[57]
NSP5 is encoded by genome segment 11 of rotavirus A. In virus-infected cells NSP5 accumulates in the viroplasm.[58]
NSP6 is a nucleic acid binding protein[59] and is encoded by gene 11 from an out-of-phase open reading frame.[60]
| RNA Segment (Gene) | Size (base pairs) | Protein | UniProt | Molecular weight kDa | Location | Copies per particle | Function |
|---|---|---|---|---|---|---|---|
| 1 | 3302 | VP1 | P22678 | 125 | At the vertices of the core | 12 | RNA-dependent RNA polymerase |
| 2 | 2690 | VP2 | A2T3R5 | 102 | Forms inner shell of the core | 120 | RNA binding |
| 3 | 2591 | VP3 | A2T3S5 | 88 | At the vertices of the core | 12 | methyltransferase mRNA capping enzyme |
| 4 | 2362 | VP4 | A2T3T2 | 87 | Surface spike | 180 | Cell attachment, virulence |
| 5 | 1611 | NSP1 | Q99FX5 | 59 | Nonstructural | 0 | 5'RNA binding, interferon antagonist |
| 6 | 1356 | VP6 | Q6LE89 | 45 | Inner Capsid | 780 | Structural and species-specific antigen |
| 7 | 1104 | NSP3 | P03536 | 37 | Nonstructural | 0 | Enhances viral mRNA activity and shut-offs cellular protein synthesis |
| 8 | 1059 | NSP2 | A2T3P0 | 35 | Nonstructural | 0 | NTPase involved in RNA packaging |
| 9 | 1062 | VP71 VP72 | P03533 | 38 and 34 | Surface | 780 | Structural and neutralisation antigen |
| 10 | 751 | NSP4 | P04512 | 20 | Nonstructural | 0 | Viroporin (enterotoxin) |
| 11 | 667 | NSP5 NSP6 | A2T3Q9 P11203 | 22 | Nonstructural | 0 | ssRNA and dsRNA binding modulator of NSP2, phosphoprotein |
This table is based on the simian rotavirus strain SA11. RNA-protein coding assignments differ in some strains.
Replication

- Attachment of the virus to the host cells, which is mediated by VP4 and VP7
- Penetration of the cell by the virus and uncoating of the viral capsid
- Plus strand ssRNA synthesis (this acts as the mRNA) synthesis, which is mediated by VP1, VP3 and VP2
- Formation of the viroplasm, viral RNA packaging and minus strand RNA synthesis and formation of the double-layered virus particles
- Virus particle maturation and release of progeny virions
The attachment of the virus to the host cell is initiated by VP4, which attaches to molecules, called glycans, on the surface of the cell.[33] The virus enters cells by receptor mediated endocytosis and form a vesicle known as an endosome. Proteins in the third layer (VP7 and the VP4 spike) disrupt the membrane of the endosome, creating a difference in the calcium concentration. This causes the breakdown of VP7 trimers into single protein subunits, leaving the VP2 and VP6 protein coats around the viral dsRNA, forming a double-layered particle (DLP).[62]
The eleven dsRNA strands remain within the protection of the two protein shells and the viral RNA-dependent RNA polymerase creates mRNA transcripts of the double-stranded viral genome. By remaining in the core, the viral RNA evades innate host immune responses including RNA interference that are triggered by the presence of double-stranded RNA.[63]
During the infection, rotaviruses produce mRNA for both protein biosynthesis and gene replication. Most of the rotavirus proteins accumulate in viroplasm, where the RNA is replicated and the DLPs are assembled. In the viroplasm the positive sense viral RNAs that are used as templates for the synthesis of viral genomic dsRNA are protected from siRNA-induced RNase degradation.[64] Viroplasm is formed around the cell nucleus as early as two hours after virus infection, and consists of viral factories thought to be made by two viral nonstructural proteins: NSP5 and NSP2. Inhibition of NSP5 by RNA interference in vitro results in a sharp decrease in rotavirus replication. The DLPs migrate to the endoplasmic reticulum where they obtain their third, outer layer (formed by VP7 and VP4). The progeny viruses are released from the cell by lysis.[44][65][66]
Transmission

Rotaviruses are transmitted by the faecal–oral route, via contact with contaminated hands, surfaces and objects,[67] and possibly by the respiratory route.[68] Viral diarrhoea is highly contagious. The faeces of an infected person can contain more than 10 trillion infectious particles per gram;[48] fewer than 100 of these are required to transmit infection to another person.[3]
Rotaviruses are stable in the environment and have been found in estuary samples at levels up to 1–5 infectious particles per US gallon. The viruses survive between 9 and 19 days.[69] Sanitary measures adequate for eliminating bacteria and parasites seem to be ineffective in control of rotavirus, as the incidence of rotavirus infection in countries with high and low health standards is similar.[68]
Signs and symptoms
Rotaviral enteritis is a mild to severe disease characterised by nausea, vomiting, watery diarrhoea and low-grade fever. Once a child is infected by the virus, there is an incubation period of about two days before symptoms appear.[70] The period of illness is acute. Symptoms often start with vomiting followed by four to eight days of profuse diarrhoea. Dehydration is more common in rotavirus infection than in most of those caused by bacterial pathogens, and is the most common cause of death related to rotavirus infection.[71]
Rotavirus infections can occur throughout life: the first usually produces symptoms, but subsequent infections are typically mild or asymptomatic,[72][48] as the immune system provides some protection.[73] Consequently, symptomatic infection rates are highest in children under two years of age and decrease progressively towards 45 years of age.[74] The most severe symptoms tend to occur in children six months to two years of age, the elderly, and those with immunodeficiency. Due to immunity acquired in childhood, most adults are not susceptible to rotavirus; gastroenteritis in adults usually has a cause other than rotavirus, but asymptomatic infections in adults may maintain the transmission of infection in the community.[75] There is some evidence to suggest blood group can impact on the susceptibility to infection by rotaviruses.[76]
Disease mechanisms

Rotaviruses replicate mainly in the gut,[77] and infect enterocytes of the villi of the small intestine, leading to structural and functional changes of the epithelium.[78] There is evidence in humans, and particularly in animal models of extraintestinal dissemination of infectious virus to other organs and macrophages.[79]
The diarrhoea is caused by multiple activities of the virus.[80] Malabsorption occurs because of the destruction of gut cells called enterocytes. The toxic rotavirus protein NSP4 induces age- and calcium ion-dependent chloride secretion, disrupts SGLT1 (sodium/glucose cotransporter 2) transporter-mediated reabsorption of water, apparently reduces activity of brush-border membrane disaccharidases, and activates the calcium ion-dependent secretory reflexes of the enteric nervous system.[56] The elevated concentrations of calcium ions in the cytosol (which are required for the assembly of the progeny viruses) is achieved by NSP4 acting as a viroporin. This increase in calcium ions leads to autophagy (self destruction) of the infected enterocytes.[81]
NSP4 is also secreted. This extracellular form, which is modified by protease enzymes in the gut, is an enterotoxin which acts on uninfected cells via integrin receptors, which in turn cause and increase in intracellular calcium ion concentrations, secretory diarrhoea and autophagy.[82]
The vomiting, which is a characteristic of rotaviral enteritis, is caused by the virus infecting the enterochromaffin cells on the lining of the digestive tract. The infection stimulates the production of 5' hydroxytryptamine (serotonin). This activates vagal afferent nerves, which in turn activates the cells of the brain stem that control the vomiting reflex.[83]
Healthy enterocytes secrete lactase into the small intestine; milk intolerance due to lactase deficiency is a symptom of rotavirus infection,[84] which can persist for weeks.[85] A recurrence of mild diarrhoea often follows the reintroduction of milk into the child's diet, due to bacterial fermentation of the disaccharide lactose in the gut.[86]
Immune responses
Specific responses
Rotaviruses elicit both B and T cell immune responses. Antibodies to the rotavirus VP4 and VP7 proteins neutralise viral infectivity in vitro and in vivo.[87] Specific antibodies of the classes IgM, IgA and IgG are produced, which have been shown to protect against rotavirus infection by the passive transfer of the antibodies in other animals.[88] Maternal trans-placental IgG might play a role in the protection neonates from rotavirus infections, but on the other hand might reduce vaccine efficacy.[89]
Innate responses
Following infection by rotaviruses there is a rapid innate immune response involving types I and III interferons and other cytokines (particularly Th1 and Th2)[90] which inhibit the replication of the virus and recruit macrophages and natural killer cells to the rotavirus infected cells.[91] The rotavirus dsRNA activates pattern recognition receptors such toll-like receptors that stimulate the production of interferons.[92] The rotavirus protein NSP1 counteracts the effects of type 1 interferons by suppressing the activity of the interferon regulatory proteins IRF3, IRF5 and IRF7.[92]
Markers of protection
The levels of IgG and IgA in the blood and IgA in the gut correlate with protection from infection.[93] Rotavirus specific serum IgG and IgA at high titres (e.g. >1:200) have been claimed to be protective and there is a significant correlation between IgA titres and rotavirus vaccine efficacy.[94]
Diagnosis and detection
Diagnosis of infection with a rotavirus normally follows diagnosis of gastroenteritis as the cause of severe diarrhoea. Most children admitted to hospital with gastroenteritis are tested for rotavirus.[95][96]
Specific diagnosis of infection with rotavirus is made by finding the virus in the child's stool by enzyme immunoassay. There are several licensed test kits on the market which are sensitive, specific and detect all serotypes of rotavirus.[97] Other methods, such as electron microscopy and PCR (polymerase chain reaction), are used in research laboratories.[98] Reverse transcription-polymerase chain reaction (RT-PCR) can detect and identify all species and serotypes of human rotaviruses.[99]
Treatment and prognosis
Treatment of acute rotavirus infection is nonspecific and involves management of symptoms and, most importantly, management of dehydration.[13] If untreated, children can die from the resulting severe dehydration.[100] Depending on the severity of diarrhoea, treatment consists of oral rehydration therapy, during which the child is given extra water to drink that contains specific amounts of salt and sugar.[101] In 2004, the World Health Organisation (WHO) and UNICEF recommended the use of low-osmolarity oral rehydration solution and zinc supplementation as a two-pronged treatment of acute diarrhoea.[102] Some infections are serious enough to warrant hospitalisation where fluids are given by intravenous therapy or nasogastric intubation, and the child's electrolytes and blood sugar are monitored.[95] Rotavirus infections rarely cause other complications and for a well managed child the prognosis is excellent.[103] Probiotics have been shown to reduce the duration of rotavirus diarrhoea,[104] and according to the European Society for Pediatric Gastroenterology "effective interventions include administration of specific probiotics such as Lactobacillus rhamnosus or Saccharomyces boulardii, diosmectite or racecadotril."[105]
Prevention
Rotaviruses are highly contagious and cannot be treated with antibiotics or other drugs. Because improved sanitation does not decrease the prevalence of rotaviral disease, and the rate of hospitalisations remains high despite the use of oral rehydrating medicines, the primary public health intervention is vaccination.[2] In 1998, a rotavirus vaccine was licensed for use in the United States. Clinical trials in the United States, Finland, and Venezuela had found it to be 80–100% effective at preventing severe diarrhoea caused by rotavirus A, and researchers had detected no statistically significant serious adverse effects.[106][107] The manufacturer, however, withdrew it from the market in 1999, after it was discovered that the vaccine may have contributed to an increased risk for intussusception, a type of bowel obstruction, in one of every 12,000 vaccinated infants.[108] The experience provoked intense debate about the relative risks and benefits of a rotavirus vaccine.[109]
In 2006, two new vaccines against rotavirus A infection were shown to be safe and effective in children,[110] and in 2009, the WHO recommended that rotavirus vaccine be included in all national immunisation programmes.[111]
The incidence and severity of rotavirus infections has declined significantly in countries that have acted on this recommendation.[14][15][16] A 2014 review of available clinical trial data from countries routinely using rotavirus vaccines in their national immunisation programs found that rotavirus vaccines have reduced rotavirus hospitalisations by 49–92% and all cause diarrhoea hospitalisations by 17–55%.[112] In Mexico, which in 2006 was among the first countries in the world to introduce rotavirus vaccine, diarrhoeal disease death rates dropped during the 2009 rotavirus season by more than 65 percent among children age two and under.[113] In Nicaragua, which in 2006 became the first developing country to introduce a rotavirus vaccine, severe rotavirus infections were reduced by 40 percent and emergency room visits by a half.[114] In the United States, rotavirus vaccination since 2006 has led to drops in rotavirus-related hospitalisations by as much as 86 percent.[115] The vaccines may also have prevented illness in non-vaccinated children by limiting the number of circulating infections.[115][116] In developing countries in Africa and Asia, where the majority of rotavirus deaths occur, a large number of safety and efficacy trials as well as recent post-introduction impact and effectiveness studies of Rotarix and RotaTeq have found that vaccines dramatically reduced severe disease among infants.[16][117][118][119] In September 2013, the vaccine was offered to all children in the UK, aged between two and three months, and it is expected to halve the cases of severe infection and reduce the number of children admitted to hospital because of the infection by 70 percent.[120] In Europe, hospitalisation rates following infection by rotaviruses have decreased by 65% to 84% following the introduction of the vaccine.[121] Globally, vaccination has reduced hospital admissions and emergency department visits by a median of 67%.[122]
Rotavirus vaccines are licensed in over 100 countries, and more than 80 countries have introduced routine rotavirus vaccination, almost half with the support of the GAVI vaccine alliance.[123] To make rotavirus vaccines available, accessible, and affordable in all countries—particularly low- and middle-income countries in Africa and Asia where the majority of rotavirus deaths occur, PATH (formerly Program for Appropriate Technology in Health), the WHO, the U.S. Centers for Disease Control and Prevention, and GAVI have partnered with research institutions and governments to generate and disseminate evidence, lower prices, and accelerate introduction.[124]
The vaccine may prevent type 1 diabetes.[125][126]
Epidemiology
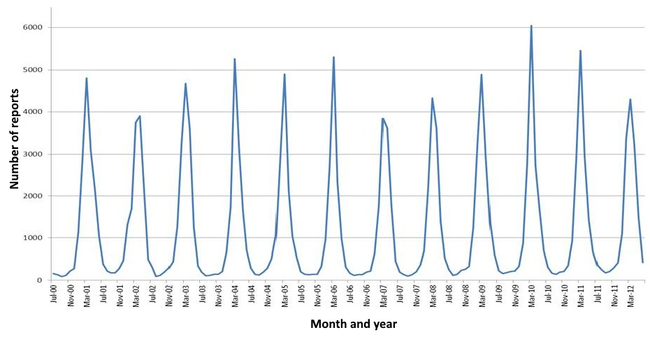
Rotavirus A, which accounts for more than 90% of rotavirus gastroenteritis in humans,[4] is endemic worldwide. Each year rotaviruses cause millions of cases of diarrhoea in developing countries, almost 2 million of which result in hospitalisation.[7] In 2019, an estimated 151,714 children younger than five died from rotavirus infections, 90 percent of whom were in developing countries.[9] Almost every child has been infected with rotaviruses by age five.[2][127] Rotaviruses are the leading single cause of severe diarrhoea among infants and children, is responsible for about a third of the cases requiring hospitalisation,[11] and causes 37% of deaths attributable to diarrhoea and 5% of all deaths in children younger than five.[128] Boys are twice as likely as girls to be admitted to hospital for rotavirus infections.[129][130] In the pre-vaccination era, rotavirus infections occurred primarily during cool, dry seasons.[131][132] The number attributable to food contamination is unknown.[133]
Outbreaks of rotavirus A diarrhoea are common among hospitalised infants, young children attending day care centres, and elderly people in nursing homes.[75][134] An outbreak caused by contaminated municipal water occurred in Colorado in 1981.[135] During 2005, the largest recorded epidemic of diarrhoea occurred in Nicaragua. This unusually large and severe outbreak was associated with mutations in the rotavirus A genome, possibly helping the virus escape the prevalent immunity in the population.[136] A similar large outbreak occurred in Brazil in 1977.[137]
Rotavirus B, also called adult diarrhoea rotavirus or ADRV, has caused major epidemics of severe diarrhoea affecting thousands of people of all ages in China. These epidemics occurred as a result of sewage contamination of drinking water.[138][139] Rotavirus B infections also occurred in India in 1998; the causative strain was named CAL. Unlike ADRV, the CAL strain is endemic.[140][141] To date, epidemics caused by rotavirus B have been confined to mainland China, and surveys indicate a lack of immunity to this species in the United States.[142] Rotavirus C has been associated with rare and sporadic cases of diarrhoea in children, and small outbreaks have occurred in families.[143]
- The seasonal variation of rotavirus A infections in England: rates of infection peak during the winter months.[144]
- Preventable child deaths from rotavirus vaccination, 2016. Annual number of preventable deaths in children under five years old from rotavirus if full coverage of the rotavirus vaccine was achieved.[145]
Other animals
Rotaviruses infect the young of many species of animals and they are a major cause of diarrhoea in wild and reared animals worldwide.[8] As a pathogen of livestock, notably in young calves and piglets, rotaviruses cause economic loss to farmers because of costs of treatment associated with high morbidity and mortality rates.[146] These rotaviruses are a potential reservoir for genetic exchange with human rotaviruses.[146] There is evidence that animal rotaviruses can infect humans, either by direct transmission of the virus or by contributing one or several RNA segments to reassortants with human strains.[147][148][149]
History

In 1943, Jacob Light and Horace Hodes proved that a filterable agent in the faeces of children with infectious diarrhoea also caused scours (livestock diarrhoea) in cattle.[150] Three decades later, preserved samples of the agent were shown to be rotavirus.[151] In the intervening years, a virus in mice[152] was shown to be related to the virus causing scours.[153] In 1973, Ruth Bishop and colleagues described related viruses found in children with gastroenteritis.[5]
In 1974, Thomas Henry Flewett suggested the name rotavirus after observing that, when viewed through an electron microscope, a rotavirus particle looks like a wheel (rota in Latin)[154][155] the name was officially recognised by the International Committee on Taxonomy of Viruses four years later.[156] In 1976, related viruses were described in several other species of animals.[153] These viruses, all causing acute gastroenteritis, were recognised as a collective pathogen affecting humans and other animals worldwide.[154] Rotavirus serotypes were first described in 1980,[157] and in the following year, rotaviruses from humans were first grown in cell cultures derived from monkey kidneys, by adding trypsin (an enzyme found in the duodenum of mammals and now known to be essential for rotavirus to replicate) to the culture medium.[158] The ability to grow rotaviruses in culture accelerated the pace of research, and by the mid-1980s the first candidate vaccines were being evaluated.[159]
References
External links
 Medicine portal
Medicine portal Viruses portal
Viruses portal Media related to Rotavirus at Wikimedia Commons
Media related to Rotavirus at Wikimedia Commons