| Babel user information | ||||||||
|---|---|---|---|---|---|---|---|---|
Template:User Scientific Linux | ||||||||
| Users by language |

User:Bci2/Tabs
User:Bci2/box-header

Physics (from Greek φυσική (ἐπιστήμη), i.e. "knowledge of nature", from φύσις, physis "nature"), is the natural science that involves the study of matter (anything that has mass and occupies space) and its motion (movement from place to another) through space and time, light and its propagation, along with related concepts such as energy and force. More broadly, it is the general analysis of nature, conducted in order to understand how the universe behaves.
Physics is one of the oldest academic disciplines, perhaps the oldest through its inclusion of astronomy. Evidence exists that the earliest civilizations dating back to beyond 3000 BCE, such as the Sumerians, Ancient Egyptians, and the Indus Valley Civilization, all had a predictive knowledge and a very basic understanding of the motions of the Sun, Moon, and stars. Although originally part of other physical sciences and mathematics, Physics emerged to become a unique modern science during the Scientific Revolution of the 16th century.
Physics is both significant and influential, in part because advances in its understanding have often translated into new technologies, but also because new ideas in physics often resonate with other sciences, mathematics, and philosophy. For example, advances in the understanding of electromagnetism or nuclear physics led directly to the development of new products which have dramatically transformed modern-day society (e.g., television, computers, domestic appliances, atomic power, and nuclear weapons); advances in thermodynamics led to the development of motorized transport and advances in aviation engineering; and advances in mechanics inspired the development of calculus.
Physics also has philosophical implications. It can be historically traced back to ancient Greek philosophy. From Thales' first attempt to characterize matter to Democritus' deduction that matter ought to reduce to an invariant state, the Ptolemaic astronomy of a crystalline firmament, and Aristotle's book Physics, different Greek philosophers advanced their own theories of nature. Well into the 18th century, physics was known as "Natural philosophy". By the 19th century, physics was realized as a positive science and a distinct discipline separate from philosophy and the other sciences. Physics, as with the rest of science, relies on philosophy of science to give an adequate description of the scientific method.User:Bci2/box-footer
User:Bci2/box-header
 |
| National Superconducting Cyclotron Laboratory |
The National Superconducting Cyclotron Laboratory (NSCL) is a United States laboratory that focusses on rare isotope research. Established in 1963, it is located on the campus of Michigan State University. Funded primarily by the National Science Foundation, the NSCL operates two superconducting cyclotrons. In December 2008, the Department of Energy announced that the NSCL would host the Faculty for Rare Isotope Beams. The NSCL employs 300 people, and awards ~10% of the United States' nuclear science doctorates.User:Bci2/box-footer
Consider also that many sources define Metamathematics as the branch of mathematics (not logic!) that deals with "the logic and consistency of mathematical proofs, formulas, and equations". Thus, a major part of metamathemtics deals with metatheorems, that is "theorems about theorems", and meta-propositions about propositions, and so on. On the other hand, metalogic is defined in Wikipedia as: "the study of the metatheory of logic." (i.e., not mathematics). To sum up, whereas metamathematics is the study of metatheories in mathematics, Metalogic is the study of the metatheories of logic. Therefore, logically, unless one is willing to say that mathematics and logic are identical or even equivalent--which they are not-- metamathematics and metalogic are not identical, but substantially different. Moreover, as there is also mathematical logic, one has to conceed there is partial overlap between the two fields of mathematics and logic, but such an overlap is not up to either logical identity or mathematical equivalence. Many mathematical logicians would like to think, or define, mathematical logic as comprising the entire field of metamathematics, which it currently does not, but it may have been the case 50 years or a century ago.
Serious metamathematical reflection began with the work of Gottlob Frege, especially his Begriffsschrift. David Hilbert was the first to invoke the term "metamathematics" with regularity (see Hilbert's program). In his hands, it meant something akin to contemporary proof theory. Another important contemporary branch is model theory. Other leading figures in the field include Bertrand Russell, Thoralf Skolem, Emil Post, Alonzo Church, Stephen Kleene, Willard Quine, Paul Benacerraf, Hilary Putnam, Gregory Chaitin, and most important, Alfred Tarski and Kurt Gödel. In particular, Gödel's proof that, given any finite number of axioms for Peano arithmetic, there will be true statements about that arithmetic that cannot be proved from those axioms, a result known as the incompleteness theorem, is arguably the greatest achievement of metamathematics and the philosophy of mathematics to date.
The List of articles initiated and created by Bci2 on Wikipedia
- American-Romanian Academy of Arts and Sciences
- European CNRS Franco-Romanian Associated Laboratories
- Higher dimensional algebra
- Theodor V. Ionescu
- R-algebroid
- Double groupoid
- Society for Mathematical Biology
- Complex system biology
- George Karreman
- Nicolas Rashevsky
- Molecular models of DNA
- Florentina Mosora
- Esquisse d'un Programme
- VCD
- Fraţii Buzeşti High School
- 2D-FT NMRI and Spectroscopy
Other Major contributions to Wikipedia articles
- George Emil Palade
- Robert Rosen
- Nicolae Popescu
- List of important publications in biology
- Systems biology
- Mathematical and theoretical biology
- Quark
- Tannaka–Krein duality
- Nuclear magnetic resonance
- Category theory
- Category theory
- Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid
- *FT-NIRS (FT-NIR) Spectroscopy
See also
References
References
- Alfred Whitehead, and Bertrand Russell. Principia Mathematica, 3 vols, Cambridge University Press, 1910, 1912, and 1913. Second edition, 1925 (Vol. 1), 1927 (Vols 2, 3). Abridged as Principia Mathematica to *56, Cambridge University Press, 1962.
- I. J. Good. "A Note on Richard's Paradox". Mind, New Series, Vol. 75, No. 299 (Jul., 1966), p. 431. JStor]
- Jules Richard, Les Principes des Mathématiques et le Problème des Ensembles, Revue Générale des Sciences Pures et Appliquées (1905); translated in Heijenoort J. van (ed.), Source Book in Mathematical Logic 1879-1931 (Cambridge, Mass., 1964).
- Douglas Hofstadter, 1980. Gödel, Escher, Bach. Vintage Books. Aimed at laypeople.
- Stephen Cole Kleene, 1952. Introduction to Metamathematics. North Holland. Aimed at mathematicians.
Categories
See also
References
- Douglas Hofstadter, 1980. Gödel, Escher, Bach. Vintage Books. Aimed at laypeople.
- Stephen Cole Kleene, 1952. Introduction to Metamathematics. North Holland. Aimed at mathematicians.
Metalogic
http://en.wikipedia.org/wiki/Metalogic
Metalogic is the study of the metatheory of logic. While logic is the study of the manner in which logical systems can be used to decide the correctness of arguments, metalogic studies the properties of the logical systems themselves.[1] According to Geoffrey Hunter, while logic concerns itself with the "truths of logic," metalogic concerns itself with the theory of "sentences used to express truths of logic."[2]
The basic objects of study in metalogic are formal languages, formal systems, and their interpretations. The study of interpretation of formal systems is the branch of mathematical logic known as model theory, while the study of deductive apparatus is the branch known as proof theory.
History
Metalogical questions have been asked since the time of Aristotle. However, it was only with the rise of formal languages in the late 19th and early 20th century that investigations into the foundations of logic began to flourish. In 1904, David Hilbert observed that in investigating the foundations of mathematics that logical notions are presupposed, and therefore a simultaneous account of metalogical and metamathematical principles was required. Today, metalogic and metamathematics are largely synonymous with each other, and both have been substantially subsumed by mathematical logic in academia.
Important distinctions in metalogic
Metalanguage-Object language
In metalogic, formal languages are sometimes called object languages. The language used to make statements about an object language is called a metalanguage. This distinction is a key difference between logic and metalogic. While logic deals with proofs in a formal system, expressed in some formal language, metalogic deals with proofs about a formal system which are expressed in a metalanguage about some object language.
Syntax-semantics
In metalogic, 'syntax' has to do with formal languages or formal systems without regard to any interpretation of them, whereas, 'semantics' has to do with interpretations of formal languages. The term 'syntactic' has a slightly wider scope than 'proof-theoretic', since it may be applied to properties of formal languages without any deductive systems, as well as to formal systems. 'Semantic' is synonymous with 'model-theoretic'.
Use-mention
In metalogic, the words 'use' and 'mention', in both their noun and verb forms, take on a technical sense in order to identify an important distinction.[2] The use–mention distinction (sometimes referred to as the words-as-words distinction) is the distinction between using a word (or phrase) and mentioning it. Usually it is indicated that an expression is being mentioned rather than used by enclosing it in quotation marks, printing it in italics, or setting the expression by itself on a line. The enclosing in quotes of an expression gives us the name of an expression, for example:
- 'Metalogic' is the name of this article.
- This article is about metalogic.
Type-token
The type-token distinction is a distinction in metalogic, that separates an abstract concept from the objects which are particular instances of the concept. For example, the particular bicycle in your garage is a token of the type of thing known as "The bicycle." Whereas, the bicycle in your garage is in a particular place at a particular time, that is not true of "the bicycle" as used in the sentence: "The bicycle has become more popular recently." This distinction is used to clarify the meaning of symbols of formal languages.
Overview
Formal language
A formal language is an organized set of symbols the essential feature of which is that it can be precisely defined in terms of just the shapes and locations of those symbols. Such a language can be defined, then, without any reference to any meanings of any of its expressions; it can exist before any formal interpretation is assigned to it -- that is, before it has any meaning. First order logic is expressed in some formal language. A formal grammar determines which symbols and sets of symbols are formulas in a formal language.
A formal language can be defined formally as a set A of strings (finite sequences) on a fixed alphabet α. Some authors, including Carnap, define the language as the ordered pair <α, A>.[3] Carnap also requires that each element of α must occur in at least one string in A.
Formal grammar
A formal grammar (also called formation rules) is a precise description of a the well-formed formulas of a formal language. It is synonymous with the set of strings over the alphabet of the formal language which constitute well formed formulas. However, it does not describe their semantics (i.e. what they mean).
Formal systems
A formal system (also called a logical calculus, or a logical system) consists of a formal language together with a deductive apparatus (also called a deductive system). The deductive apparatus may consist of a set of transformation rules (also called inference rules) or a set of axioms, or have both. A formal system is used to derive one expression from one or more other expressions.
A formal system can be formally defined as an ordered triple <α,,d>, where d is the relation of direct derivability. This relation is understood in a comprehensive sense such that the primitive sentences of the formal system are taken as directly derivable from the empty set of sentences. Direct derivability is a relation between a sentence and a finite, possibly empty set of sentences. Axioms are laid down in such a way that every first place member of d is a member of and every second place member is a finite subset of .
It is also possible to define a formal system using only the relation d. In this way we can omit , and α in the definitions of interpreted formal language, and interpreted formal system. However, this method can be more difficult to understand and work with.[3]
Formal proofs
A formal proof is a sequences of well-formed formulas of a formal language, the last one of which is a theorem of a formal system. The theorem is a syntactic consequence of all the well formed formulae preceding it in the proof. For a well formed formula to qualify as part of a proof, it must be the result of applying a rule of the deductive apparatus of some formal system to the previous well formed formulae in the proof sequence.
Formal interpretations
A formal interpretation of a formal system is the assignment of meanings, to the symbols, and truth-values to the sentences of the formal system. The study of formal interpretations is called Formal semantics. Giving an interpretation is synonymous with constructing a model.
Results in metalogic
Results in metalogic consist of such things as formal proofs demonstrating the consistency, completeness, and decidability of particular formal systems.
Major results in metalogic include:
- Proof of the uncountability of the set of all subsets of the set of natural numbers (Cantor's theorem 1891)
- Löwenheim-Skolem theorem (Leopold Löwenheim 1915 and Thoralf Skolem 1919)
- Proof of the consistency of truth-functional propositional logic (Emil Post 1920)
- Proof of the semantic completeness of truth-functional propositional logic (Paul Bernays 1918)[4],(Emil Post 1920)[2]
- Proof of the syntactic completeness of truth-functional propositional logic (Emil Post 1920)[2]
- Proof of the decidability of truth-functional propositional logic (Emil Post 1920)[2]
- Proof of the consistency of first order monadic predicate logic (Leopold Löwenheim 1915)
- Proof of the semantic completeness of first order monadic predicate logic (Leopold Löwenheim 1915)
- Proof of the decidability of first order monadic predicate logic (Leopold Löwenheim 1915)
- Proof of the semantic completeness of first order predicate logic (Gödel's completeness theorem 1930)
- Proof of the consistency of first order predicate logic (David Hilbert and Wilhelm Ackermann 1928)
- Proof of the semantic completeness of first order predicate logic (Kurt Gödel 1930)
- Proof of the undecidability of first order predicate logic (Alonzo Church 1936)
- Gödel's first incompleteness theorem 1931
- Gödel's second incompleteness theorem 1931
See also
References

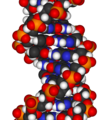
Deoxyribonucleic acid (DNA) is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms and some viruses. The main role of DNA molecules is the long-term storage of information. DNA is often compared to a set of blueprints or a recipe, or a code, since it contains the instructions needed to construct other components of cells, such as proteins and RNA molecules. The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in regulating the use of this genetic information.
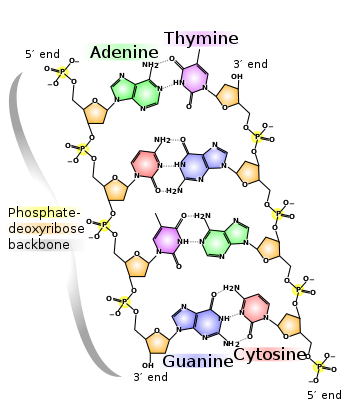
Chemically, DNA consists of two long polymers of simple units called nucleotides, with backbones made of sugars and phosphate groups joined by ester bonds. These two strands run in opposite directions to each other and are therefore anti-parallel. Attached to each sugar is one of four types of molecules called bases. It is the sequence of these four bases along the backbone that encodes information. This information is read using the genetic code, which specifies the sequence of the amino acids within proteins. The code is read by copying stretches of DNA into the related nucleic acid RNA, in a process called transcription.
Within cells, DNA is organized into structures called chromosomes. These chromosomes are duplicated before cells divide, in a process called DNA replication. Eukaryotic organisms (animals, plants, fungi, and protists) store their DNA inside the cell nucleus, while in prokaryotes (bacteria and archaea) it is found in the cell's cytoplasm. Within the chromosomes, chromatin proteins such as histones compact and organize DNA. These compact structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed.
Properties
DNA is a long polymer made from repeating units called nucleotides.[1][2][3] The DNA chain is 22 to 26 Ångströms wide (2.2 to 2.6 nanometres), and one nucleotide unit is 3.3 Å (0.33 nm) long.[4] Although each individual repeating unit is very small, DNA polymers can be very large molecules containing millions of nucleotides. For instance, the largest human chromosome, chromosome number 1, is approximately 220 million base pairs long.[5]
In living organisms, DNA does not usually exist as a single molecule, but instead as a tightly associated pair of molecules.[6][7] These two long strands entwine like vines, in the shape of a double helix. The nucleotide repeats contain both the segment of the backbone of the molecule, which holds the chain together, and a base, which interacts with the other DNA strand in the helix. In general, a base linked to a sugar is called a nucleoside and a base linked to a sugar and one or more phosphate groups is called a nucleotide. If multiple nucleotides are linked together, as in DNA, this polymer is called a polynucleotide.[8]
The backbone of the DNA strand is made from alternating phosphate and sugar residues.[9] The sugar in DNA is 2-deoxyribose, which is a pentose (five-carbon) sugar. The sugars are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These asymmetric bonds mean a strand of DNA has a direction. In a double helix the direction of the nucleotides in one strand is opposite to their direction in the other strand. This arrangement of DNA strands is called antiparallel. The asymmetric ends of DNA strands are referred to as the 5′ (five prime) and 3′ (three prime) ends, with the 5' end being that with a terminal phosphate group and the 3' end that with a terminal hydroxyl group. One of the major differences between DNA and RNA is the sugar, with 2-deoxyribose being replaced by the alternative pentose sugar ribose in RNA.[7]
The DNA double helix is stabilized by hydrogen bonds between the bases attached to the two strands. The four bases found in DNA are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). These four bases are attached to the sugar/phosphate to form the complete nucleotide, as shown for adenosine monophosphate.
These bases are classified into two types; adenine and guanine are fused five- and six-membered heterocyclic compounds called purines, while cytosine and thymine are six-membered rings called pyrimidines.[7] A fifth pyrimidine base, called uracil (U), usually takes the place of thymine in RNA and differs from thymine by lacking a methyl group on its ring. Uracil is not usually found in DNA, occurring only as a breakdown product of cytosine.
Grooves

Normally, the double helix is a right-handed spiral. As the DNA strands wind around each other, they leave gaps between each set of phosphate backbones, revealing the sides of the bases inside (see animation). There are two of these grooves twisting around the surface of the double helix: one groove, the major groove, is 22 Å wide and the other, the minor groove, is 12 Å wide.[11] The narrowness of the minor groove means that the edges of the bases are more accessible in the major groove. As a result, proteins like transcription factors that can bind to specific sequences in double-stranded DNA usually make contacts to the sides of the bases exposed in the major groove.[12] This situation varies in unusual conformations of DNA within the cell (see below), but the major and minor grooves are always named to reflect the differences in size that would be seen if the DNA is twisted back into the ordinary B form.
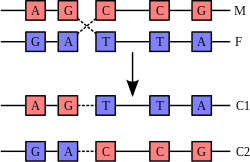
Base pairing
Each type of base on one strand forms a bond with just one type of base on the other strand. This is called complementary base pairing. Here, purines form hydrogen bonds to pyrimidines, with A bonding only to T, and C bonding only to G. This arrangement of two nucleotides binding together across the double helix is called a base pair. As hydrogen bonds are not covalent, they can be broken and rejoined relatively easily. The two strands of DNA in a double helix can therefore be pulled apart like a zipper, either by a mechanical force or high temperature.[13] As a result of this complementarity, all the information in the double-stranded sequence of a DNA helix is duplicated on each strand, which is vital in DNA replication. Indeed, this reversible and specific interaction between complementary base pairs is critical for all the functions of DNA in living organisms.[2]
 |
 |
The two types of base pairs form different numbers of hydrogen bonds, AT forming two hydrogen bonds, and GC forming three hydrogen bonds (see figures, left). DNA with high GC-content is more stable than DNA with low GC-content, but contrary to popular belief, this is not due to the extra hydrogen bond of a GC basepair but rather the contribution of stacking interactions (hydrogen bonding merely provides specificity of the pairing, not stability).[14]As a result, it is both the percentage of GC base pairs and the overall length of a DNA double helix that determine the strength of the association between the two strands of DNA. Long DNA helices with a high GC content have stronger-interacting strands, while short helices with high AT content have weaker-interacting strands.[15] In biology, parts of the DNA double helix that need to separate easily, such as the TATAAT Pribnow box in some promoters, tend to have a high AT content, making the strands easier to pull apart.[16] In the laboratory, the strength of this interaction can be measured by finding the temperature required to break the hydrogen bonds, their melting temperature (also called Tm value). When all the base pairs in a DNA double helix melt, the strands separate and exist in solution as two entirely independent molecules. These single-stranded DNA molecules have no single common shape, but some conformations are more stable than others.[17]
Sense and antisense
A DNA sequence is called "sense" if its sequence is the same as that of a messenger RNA copy that is translated into protein.[18] The sequence on the opposite strand is called the "antisense" sequence. Both sense and antisense sequences can exist on different parts of the same strand of DNA (i.e. both strands contain both sense and antisense sequences). In both prokaryotes and eukaryotes, antisense RNA sequences are produced, but the functions of these RNAs are not entirely clear.[19] One proposal is that antisense RNAs are involved in regulating gene expression through RNA-RNA base pairing.[20]
A few DNA sequences in prokaryotes and eukaryotes, and more in plasmids and viruses, blur the distinction between sense and antisense strands by having overlapping genes.[21] In these cases, some DNA sequences do double duty, encoding one protein when read along one strand, and a second protein when read in the opposite direction along the other strand. In bacteria, this overlap may be involved in the regulation of gene transcription,[22] while in viruses, overlapping genes increase the amount of information that can be encoded within the small viral genome.[23]
Supercoiling
DNA can be twisted like a rope in a process called DNA supercoiling. With DNA in its "relaxed" state, a strand usually circles the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted the strands become more tightly or more loosely wound.[24] If the DNA is twisted in the direction of the helix, this is positive supercoiling, and the bases are held more tightly together. If they are twisted in the opposite direction, this is negative supercoiling, and the bases come apart more easily. In nature, most DNA has slight negative supercoiling that is introduced by enzymes called topoisomerases.[25] These enzymes are also needed to relieve the twisting stresses introduced into DNA strands during processes such as transcription and DNA replication.[26]

Alternate DNA structures

DNA exists in many possible conformations that include A-DNA, B-DNA, and Z-DNA forms, although, only B-DNA and Z-DNA have been directly observed in organisms.[9] The conformation that DNA adopts depends on the hydration level, DNA sequence, the amount and direction of supercoiling, chemical modifications of the bases, the type and concentration of metal ions, as well as the presence of polyamines in solution[28].
The first published reports of A-DNA X-ray diffraction patterns-- and also B-DNA-- employed analyses based on Patterson transforms that provided only a limited amount of structural information for oriented fibers of DNA isolated from calf thymus.[29][30] An alternate analysis was then proposed by Wilkins et al. in 1953 for B-DNA X-ray diffraction/scattering patterns of hydrated, bacterial oriented DNA fibers and trout sperm heads in terms of squares of Bessel functions.[31] Although the `B-DNA form' is most common under the conditions found in cells,[32] it is not a well-defined conformation but a family or fuzzy set of DNA-conformations that occur at the high hydration levels present in a wide variety of living cells.[33] Their corresponding X-ray diffraction & scattering patterns are characteristic of molecular paracrystals with a significant degree of disorder (>20%)[34][35], and concomitantly the structure is not tractable using only the standard analysis.
On the other hand, the standard analysis, involving only Fourier transforms of Bessel functions[36] and DNA molecular models, is still routinely employed for the analysis of A-DNA and Z-DNA X-ray diffraction patterns.[37]
Compared to B-DNA, the A-DNA form is a wider right-handed spiral, with a shallow, wide minor groove and a narrower, deeper major groove. The A form occurs under non-physiological conditions in partially dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands, as well as in enzyme-DNA complexes.[38][39] Segments of DNA where the bases have been chemically modified by methylation may undergo a larger change in conformation and adopt the Z form. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form.[40] These unusual structures can be recognized by specific Z-DNA binding proteins and may be involved in the regulation of transcription.[41]

Quadruplex structures
At the ends of the linear chromosomes are specialized regions of DNA called telomeres. The main function of these regions is to allow the cell to replicate chromosome ends using the enzyme telomerase, as the enzymes that normally replicate DNA cannot copy the extreme 3′ ends of chromosomes.[43] These specialized chromosome caps also help protect the DNA ends, and stop the DNA repair systems in the cell from treating them as damage to be corrected.[44] In human cells, telomeres are usually lengths of single-stranded DNA containing several thousand repeats of a simple TTAGGG sequence.[45]
These guanine-rich sequences may stabilize chromosome ends by forming structures of stacked sets of four-base units, rather than the usual base pairs found in other DNA molecules. Here, four guanine bases form a flat plate and these flat four-base units then stack on top of each other, to form a stable G-quadruplex structure.[46] These structures are stabilized by hydrogen bonding between the edges of the bases and chelation of a metal ion in the centre of each four-base unit.[47] Other structures can also be formed, with the central set of four bases coming from either a single strand folded around the bases, or several different parallel strands, each contributing one base to the central structure.
In addition to these stacked structures, telomeres also form large loop structures called telomere loops, or T-loops. Here, the single-stranded DNA curls around in a long circle stabilized by telomere-binding proteins.[48] At the very end of the T-loop, the single-stranded telomere DNA is held onto a region of double-stranded DNA by the telomere strand disrupting the double-helical DNA and base pairing to one of the two strands. This triple-stranded structure is called a displacement loop or D-loop.[46]
Branched DNA
In DNA fraying occurs when non-complementary regions exist at the end of an otherwise complementary double-strand of DNA. However, branched DNA can occur if a third strand of DNA is introduced and contains adjoining regions able to hybridize with the frayed regions of the pre-existing double-strand. Although the simplest example of branched DNA involves only three strands of DNA, complexes involving additional strands and multiple branches are also possible.[49]
- A DNA structure with a single branching point.
- A DNA structure with multiple branches.
Chemical modifications
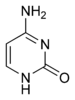 |  | 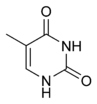 |
| cytosine | 5-methylcytosine | thymine |
Base modifications
The expression of genes is influenced by how the DNA is packaged in chromosomes, in a structure called chromatin. Base modifications can be involved in packaging, with regions that have low or no gene expression usually containing high levels of methylation of cytosine bases. For example, cytosine methylation, produces 5-methylcytosine, which is important for X-chromosome inactivation.[50] The average level of methylation varies between organisms - the worm Caenorhabditis elegans lacks cytosine methylation, while vertebrates have higher levels, with up to 1% of their DNA containing 5-methylcytosine.[51] Despite the importance of 5-methylcytosine, it can deaminate to leave a thymine base, methylated cytosines are therefore particularly prone to mutations.[52] Other base modifications include adenine methylation in bacteria and the glycosylation of uracil to produce the "J-base" in kinetoplastids.[53][54]
Damage

DNA can be damaged by many different sorts of mutagens, which change the DNA sequence. Mutagens include oxidizing agents, alkylating agents and also high-energy electromagnetic radiation such as ultraviolet light and X-rays. The type of DNA damage produced depends on the type of mutagen. For example, UV light can damage DNA by producing thymine dimers, which are cross-links between pyrimidine bases.[56] On the other hand, oxidants such as free radicals or hydrogen peroxide produce multiple forms of damage, including base modifications, particularly of guanosine, and double-strand breaks.[57] A typical human cell contains about 150,000 bases that have suffered oxidative damage.[58] Of these oxidative lesions, the most dangerous are double-strand breaks, as these are difficult to repair and can produce point mutations, insertions and deletions from the DNA sequence, as well as chromosomal translocations.[59]
Many mutagens fit into the space between two adjacent base pairs, this is called intercalating. Most intercalators are aromatic and planar molecules, and include Ethidium bromide, daunomycin, and doxorubicin. In order for an intercalator to fit between base pairs, the bases must separate, distorting the DNA strands by unwinding of the double helix. This inhibits both transcription and DNA replication, causing toxicity and mutations. As a result, DNA intercalators are often carcinogens, and Benzo[a]pyrene diol epoxide, acridines, aflatoxin and ethidium bromide are well-known examples.[60][61][62] Nevertheless, due to their ability to inhibit DNA transcription and replication, other similar toxins are also used in chemotherapy to inhibit rapidly growing cancer cells.[63]
Biological functions
DNA usually occurs as linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. The set of chromosomes in a cell makes up its genome; the human genome has approximately 3 billion base pairs of DNA arranged into 46 chromosomes.[64] The information carried by DNA is held in the sequence of pieces of DNA called genes. Transmission of genetic information in genes is achieved via complementary base pairing. For example, in transcription, when a cell uses the information in a gene, the DNA sequence is copied into a complementary RNA sequence through the attraction between the DNA and the correct RNA nucleotides. Usually, this RNA copy is then used to make a matching protein sequence in a process called translation which depends on the same interaction between RNA nucleotides. Alternatively, a cell may simply copy its genetic information in a process called DNA replication. The details of these functions are covered in other articles; here we focus on the interactions between DNA and other molecules that mediate the function of the genome.
Genes and genomes
Genomic DNA is located in the cell nucleus of eukaryotes, as well as small amounts in mitochondria and chloroplasts. In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the nucleoid.[65] The genetic information in a genome is held within genes, and the complete set of this information in an organism is called its genotype. A gene is a unit of heredity and is a region of DNA that influences a particular characteristic in an organism. Genes contain an open reading frame that can be transcribed, as well as regulatory sequences such as promoters and enhancers, which control the transcription of the open reading frame.
In many species, only a small fraction of the total sequence of the genome encodes protein. For example, only about 1.5% of the human genome consists of protein-coding exons, with over 50% of human DNA consisting of non-coding repetitive sequences.[66] The reasons for the presence of so much non-coding DNA in eukaryotic genomes and the extraordinary differences in genome size, or C-value, among species represent a long-standing puzzle known as the "C-value enigma."[67] However, DNA sequences that do not code protein may still encode functional non-coding RNA molecules, which are involved in the regulation of gene expression.[68]
Some non-coding DNA sequences play structural roles in chromosomes. Telomeres and centromeres typically contain few genes, but are important for the function and stability of chromosomes.[44][70] An abundant form of non-coding DNA in humans are pseudogenes, which are copies of genes that have been disabled by mutation.[71] These sequences are usually just molecular fossils, although they can occasionally serve as raw genetic material for the creation of new genes through the process of gene duplication and divergence.[72]
Transcription and translation
A gene is a sequence of DNA that contains genetic information and can influence the phenotype of an organism. Within a gene, the sequence of bases along a DNA strand defines a messenger RNA sequence, which then defines one or more protein sequences. The relationship between the nucleotide sequences of genes and the amino-acid sequences of proteins is determined by the rules of translation, known collectively as the genetic code. The genetic code consists of three-letter 'words' called codons formed from a sequence of three nucleotides (e.g. ACT, CAG, TTT).
In transcription, the codons of a gene are copied into messenger RNA by RNA polymerase. This RNA copy is then decoded by a ribosome that reads the RNA sequence by base-pairing the messenger RNA to transfer RNA, which carries amino acids. Since there are 4 bases in 3-letter combinations, there are 64 possible codons ( combinations). These encode the twenty standard amino acids, giving most amino acids more than one possible codon. There are also three 'stop' or 'nonsense' codons signifying the end of the coding region; these are the TAA, TGA and TAG codons.
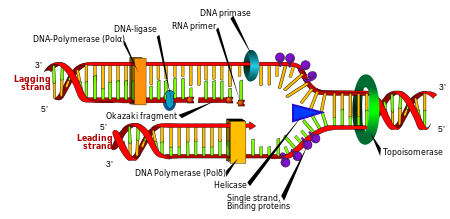
Replication
Cell division is essential for an organism to grow, but when a cell divides it must replicate the DNA in its genome so that the two daughter cells have the same genetic information as their parent. The double-stranded structure of DNA provides a simple mechanism for DNA replication. Here, the two strands are separated and then each strand's complementary DNA sequence is recreated by an enzyme called DNA polymerase. This enzyme makes the complementary strand by finding the correct base through complementary base pairing, and bonding it onto the original strand. As DNA polymerases can only extend a DNA strand in a 5′ to 3′ direction, different mechanisms are used to copy the antiparallel strands of the double helix.[73] In this way, the base on the old strand dictates which base appears on the new strand, and the cell ends up with a perfect copy of its DNA.
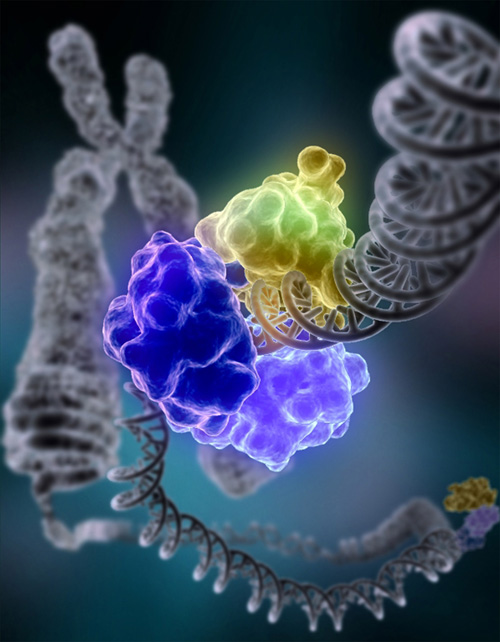
Interactions with proteins
All the functions of DNA depend on interactions with proteins. These protein interactions can be non-specific, or the protein can bind specifically to a single DNA sequence. Enzymes can also bind to DNA and of these, the polymerases that copy the DNA base sequence in transcription and DNA replication are particularly important.
DNA-binding proteins
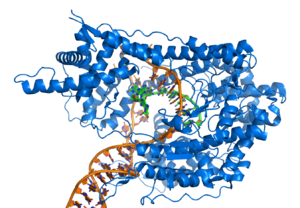
 |
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called chromatin. In eukaryotes this structure involves DNA binding to a complex of small basic proteins called histones, while in prokaryotes multiple types of proteins are involved.[74][75] The histones form a disk-shaped complex called a nucleosome, which contains two complete turns of double-stranded DNA wrapped around its surface. These non-specific interactions are formed through basic residues in the histones making ionic bonds to the acidic sugar-phosphate backbone of the DNA, and are therefore largely independent of the base sequence.[76] Chemical modifications of these basic amino acid residues include methylation, phosphorylation and acetylation.[77] These chemical changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible to transcription factors and changing the rate of transcription.[78] Other non-specific DNA-binding proteins in chromatin include the high-mobility group proteins, which bind to bent or distorted DNA.[79] These proteins are important in bending arrays of nucleosomes and arranging them into the larger structures that make up chromosomes.[80]
A distinct group of DNA-binding proteins are the DNA-binding proteins that specifically bind single-stranded DNA. In humans, replication protein A is the best-understood member of this family and is used in processes where the double helix is separated, including DNA replication, recombination and DNA repair.[81] These binding proteins seem to stabilize single-stranded DNA and protect it from forming stem-loops or being degraded by nucleases.

In contrast, other proteins have evolved to bind to particular DNA sequences. The most intensively studied of these are the various transcription factors, which are proteins that regulate transcription. Each transcription factor binds to one particular set of DNA sequences and activates or inhibits the transcription of genes that have these sequences close to their promoters. The transcription factors do this in two ways. Firstly, they can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription.[83] Alternatively, transcription factors can bind enzymes that modify the histones at the promoter; this will change the accessibility of the DNA template to the polymerase.[84]
As these DNA targets can occur throughout an organism's genome, changes in the activity of one type of transcription factor can affect thousands of genes.[85] Consequently, these proteins are often the targets of the signal transduction processes that control responses to environmental changes or cellular differentiation and development. The specificity of these transcription factors' interactions with DNA come from the proteins making multiple contacts to the edges of the DNA bases, allowing them to "read" the DNA sequence. Most of these base-interactions are made in the major groove, where the bases are most accessible.[86]

DNA-modifying enzymes
Nucleases and ligases
Nucleases are enzymes that cut DNA strands by catalyzing the hydrolysis of the phosphodiester bonds. Nucleases that hydrolyse nucleotides from the ends of DNA strands are called exonucleases, while endonucleases cut within strands. The most frequently used nucleases in molecular biology are the restriction endonucleases, which cut DNA at specific sequences. For instance, the EcoRV enzyme shown to the left recognizes the 6-base sequence 5′-GAT|ATC-3′ and makes a cut at the vertical line. In nature, these enzymes protect bacteria against phage infection by digesting the phage DNA when it enters the bacterial cell, acting as part of the restriction modification system.[88] In technology, these sequence-specific nucleases are used in molecular cloning and DNA fingerprinting.
Enzymes called DNA ligases can rejoin cut or broken DNA strands.[89] Ligases are particularly important in lagging strand DNA replication, as they join together the short segments of DNA produced at the replication fork into a complete copy of the DNA template. They are also used in DNA repair and genetic recombination.[89]
Topoisomerases and helicases
Topoisomerases are enzymes with both nuclease and ligase activity. These proteins change the amount of supercoiling in DNA. Some of these enzyme work by cutting the DNA helix and allowing one section to rotate, thereby reducing its level of supercoiling; the enzyme then seals the DNA break.[25] Other types of these enzymes are capable of cutting one DNA helix and then passing a second strand of DNA through this break, before rejoining the helix.[90] Topoisomerases are required for many processes involving DNA, such as DNA replication and transcription.[26]
Helicases are proteins that are a type of molecular motor. They use the chemical energy in nucleoside triphosphates, predominantly ATP, to break hydrogen bonds between bases and unwind the DNA double helix into single strands.[91] These enzymes are essential for most processes where enzymes need to access the DNA bases.
Polymerases
Polymerases are enzymes that synthesize polynucleotide chains from nucleoside triphosphates. The sequence of their products are copies of existing polynucleotide chains - which are called templates. These enzymes function by adding nucleotides onto the 3′ hydroxyl group of the previous nucleotide in a DNA strand. Consequently, all polymerases work in a 5′ to 3′ direction.[92] In the active site of these enzymes, the incoming nucleoside triphosphate base-pairs to the template: this allows polymerases to accurately synthesize the complementary strand of their template. Polymerases are classified according to the type of template that they use.
In DNA replication, a DNA-dependent DNA polymerase makes a copy of a DNA sequence. Accuracy is vital in this process, so many of these polymerases have a proofreading activity. Here, the polymerase recognizes the occasional mistakes in the synthesis reaction by the lack of base pairing between the mismatched nucleotides. If a mismatch is detected, a 3′ to 5′ exonuclease activity is activated and the incorrect base removed.[93] In most organisms DNA polymerases function in a large complex called the replisome that contains multiple accessory subunits, such as the DNA clamp or helicases.[94]
RNA-dependent DNA polymerases are a specialized class of polymerases that copy the sequence of an RNA strand into DNA. They include reverse transcriptase, which is a viral enzyme involved in the infection of cells by retroviruses, and telomerase, which is required for the replication of telomeres.[43][95] Telomerase is an unusual polymerase because it contains its own RNA template as part of its structure.[44]
Transcription is carried out by a DNA-dependent RNA polymerase that copies the sequence of a DNA strand into RNA. To begin transcribing a gene, the RNA polymerase binds to a sequence of DNA called a promoter and separates the DNA strands. It then copies the gene sequence into a messenger RNA transcript until it reaches a region of DNA called the terminator, where it halts and detaches from the DNA. As with human DNA-dependent DNA polymerases, RNA polymerase II, the enzyme that transcribes most of the genes in the human genome, operates as part of a large protein complex with multiple regulatory and accessory subunits.[96]
Genetic recombination
 |
 |
A DNA helix usually does not interact with other segments of DNA, and in human cells the different chromosomes even occupy separate areas in the nucleus called "chromosome territories".[98] This physical separation of different chromosomes is important for the ability of DNA to function as a stable repository for information, as one of the few times chromosomes interact is during chromosomal crossover when they recombine. Chromosomal crossover is when two DNA helices break, swap a section and then rejoin.
Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of natural selection and can be important in the rapid evolution of new proteins.[99] Genetic recombination can also be involved in DNA repair, particularly in the cell's response to double-strand breaks.[100]
The most common form of chromosomal crossover is homologous recombination, where the two chromosomes involved share very similar sequences. Non-homologous recombination can be damaging to cells, as it can produce chromosomal translocations and genetic abnormalities. The recombination reaction is catalyzed by enzymes known as recombinases, such as RAD51.[101] The first step in recombination is a double-stranded break either caused by an endonuclease or damage to the DNA.[102] A series of steps catalyzed in part by the recombinase then leads to joining of the two helices by at least one Holliday junction, in which a segment of a single strand in each helix is annealed to the complementary strand in the other helix. The Holliday junction is a tetrahedral junction structure that can be moved along the pair of chromosomes, swapping one strand for another. The recombination reaction is then halted by cleavage of the junction and re-ligation of the released DNA.[103]
Evolution
DNA contains the genetic information that allows all modern living things to function, grow and reproduce. However, it is unclear how long in the 4-billion-year history of life DNA has performed this function, as it has been proposed that the earliest forms of life may have used RNA as their genetic material.[92][104] RNA may have acted as the central part of early cell metabolism as it can both transmit genetic information and carry out catalysis as part of ribozymes.[105] This ancient RNA world where nucleic acid would have been used for both catalysis and genetics may have influenced the evolution of the current genetic code based on four nucleotide bases. This would occur since the number of unique bases in such an organism is a trade-off between a small number of bases increasing replication accuracy and a large number of bases increasing the catalytic efficiency of ribozymes.[106]
Unfortunately, there is no direct evidence of ancient genetic systems, as recovery of DNA from most fossils is impossible. This is because DNA will survive in the environment for less than one million years and slowly degrades into short fragments in solution.[107] Claims for older DNA have been made, most notably a report of the isolation of a viable bacterium from a salt crystal 250-million years old,[108] but these claims are controversial.[109][110]
Uses in technology
Genetic engineering
Methods have been developed to purify DNA from organisms, such as phenol-chloroform extraction and manipulate it in the laboratory, such as restriction digests and the polymerase chain reaction. Modern biology and biochemistry make intensive use of these techniques in recombinant DNA technology. Recombinant DNA is a man-made DNA sequence that has been assembled from other DNA sequences. They can be transformed into organisms in the form of plasmids or in the appropriate format, by using a viral vector.[111] The genetically modified organisms produced can be used to produce products such as recombinant proteins, used in medical research,[112] or be grown in agriculture.[113][114]
Forensics
Forensic scientists can use DNA in blood, semen, skin, saliva or hair found at a crime scene to identify a matching DNA of an individual, such as a perpetrator. This process is called genetic fingerprinting, or more accurately, DNA profiling. In DNA profiling, the lengths of variable sections of repetitive DNA, such as short tandem repeats and minisatellites, are compared between people. This method is usually an extremely reliable technique for identifying a matching DNA.[115] However, identification can be complicated if the scene is contaminated with DNA from several people.[116] DNA profiling was developed in 1984 by British geneticist Sir Alec Jeffreys,[117] and first used in forensic science to convict Colin Pitchfork in the 1988 Enderby murders case.[118]
People convicted of certain types of crimes may be required to provide a sample of DNA for a database. This has helped investigators solve old cases where only a DNA sample was obtained from the scene. DNA profiling can also be used to identify victims of mass casualty incidents.[119] On the other hand, many convicted people have been released from prison on the basis of DNA techniques, which were not available when a crime had originally been committed.
Bioinformatics
Bioinformatics involves the manipulation, searching, and data mining of DNA sequence data. The development of techniques to store and search DNA sequences have led to widely applied advances in computer science, especially string searching algorithms, machine learning and database theory.[120] String searching or matching algorithms, which find an occurrence of a sequence of letters inside a larger sequence of letters, were developed to search for specific sequences of nucleotides.[121] In other applications such as text editors, even simple algorithms for this problem usually suffice, but DNA sequences cause these algorithms to exhibit near-worst-case behaviour due to their small number of distinct characters. The related problem of sequence alignment aims to identify homologous sequences and locate the specific mutations that make them distinct. These techniques, especially multiple sequence alignment, are used in studying phylogenetic relationships and protein function.[122] Data sets representing entire genomes' worth of DNA sequences, such as those produced by the Human Genome Project, are difficult to use without annotations, which label the locations of genes and regulatory elements on each chromosome. Regions of DNA sequence that have the characteristic patterns associated with protein- or RNA-coding genes can be identified by gene finding algorithms, which allow researchers to predict the presence of particular gene products in an organism even before they have been isolated experimentally.[123]
DNA nanotechnology

DNA nanotechnology uses the unique molecular recognition properties of DNA and other nucleic acids to create self-assembling branched DNA complexes with useful properties. DNA is thus used as a structural material rather than as a carrier of biological information. This has led to the creation of two-dimensional periodic lattices (both tile-based as well as using the "DNA origami" method) as well as three-dimensional structures in the shapes of polyhedra. Nanomechanical devices and algorithmic self-assembly have also been demonstrated, and these DNA structures have been used to template the arrangement of other molecules such as gold nanoparticles and streptavidin proteins.
History and anthropology
Because DNA collects mutations over time, which are then inherited, it contains historical information and by comparing DNA sequences, geneticists can infer the evolutionary history of organisms, their phylogeny.[124] This field of phylogenetics is a powerful tool in evolutionary biology. If DNA sequences within a species are compared, population geneticists can learn the history of particular populations. This can be used in studies ranging from ecological genetics to anthropology; for example, DNA evidence is being used to try to identify the Ten Lost Tribes of Israel.[125][126]
DNA has also been used to look at modern family relationships, such as establishing family relationships between the descendants of Sally Hemings and Thomas Jefferson. This usage is closely related to the use of DNA in criminal investigations detailed above. Indeed, some criminal investigations have been solved when DNA from crime scenes has matched relatives of the guilty individual.[127]
History of DNA research


DNA was first isolated by the Swiss physician Friedrich Miescher who, in 1869, discovered a microscopic substance in the pus of discarded surgical bandages. As it resided in the nuclei of cells, he called it "nuclein".[128] In 1919, Phoebus Levene identified the base, sugar and phosphate nucleotide unit.[129] Levene suggested that DNA consisted of a string of nucleotide units linked together through the phosphate groups. However, Levene thought the chain was short and the bases repeated in a fixed order. In 1937 William Astbury produced the first X-ray diffraction patterns that showed that DNA had a regular structure.[130]
In 1928, Frederick Griffith discovered that traits of the "smooth" form of the Pneumococcus could be transferred to the "rough" form of the same bacteria by mixing killed "smooth" bacteria with the live "rough" form.[131] This system provided the first clear suggestion that DNA carried genetic information—the Avery-MacLeod-McCarty experiment—when Oswald Avery, along with coworkers Colin MacLeod and Maclyn McCarty, identified DNA as the transforming principle in 1943.[132] DNA's role in heredity was confirmed in 1952, when Alfred Hershey and Martha Chase in the Hershey-Chase experiment showed that DNA is the genetic material of the T2 phage.[133]
In 1953, based on X-ray diffraction images taken by Rosalind Franklin and the information that the bases were paired, James D. Watson and Francis Crick suggested what is now accepted as the first accurate model of DNA structure in the journal Nature.[6] Experimental evidence for Watson and Crick's model were published in a series of five articles in the same issue of Nature.[134] Of these, Franklin and Raymond Gosling's paper was the first publication of X-ray diffraction data that supported the Watson and Crick model;[30][135] this issue also contained an article on DNA structure by Maurice Wilkins and two of his colleagues.[31] In 1962, after Franklin's death, Watson, Crick, and Wilkins jointly received the Nobel Prize in Physiology or Medicine[136] Unfortunately, Nobel rules of the time allowed only living recipients, but a vigorous debate continues on who should receive credit for the discovery.[137]
In an influential presentation in 1957, Crick laid out the "Central Dogma" of molecular biology, which foretold the relationship between DNA, RNA, and proteins, and articulated the "adaptor hypothesis".[138] Final confirmation of the replication mechanism that was implied by the double-helical structure followed in 1958 through the Meselson-Stahl experiment.[139] Further work by Crick and coworkers showed that the genetic code was based on non-overlapping triplets of bases, called codons, allowing Har Gobind Khorana, Robert W. Holley and Marshall Warren Nirenberg to decipher the genetic code.[140] These findings represent the birth of molecular biology.
See also
- Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid
- DNA microarray
- DNA sequencing
- Paracrystal model and theory
- X-ray scattering
- Crystallography
- Genetic disorder
- Junk DNA
- Nucleic acid analogues
- Nucleic acid methods
- Nucleic acid modeling
- Nucleic acid notations
- Phosphoramidite
- Plasmid
- Polymerase chain reaction
- Proteopedia DNA
- Southern blot
- Triple-stranded DNA
Notes
Further reading
- Calladine, Chris R.; Drew, Horace R.; Luisi, Ben F. and Travers, Andrew A. Understanding DNA, Elsevier Academic Press, 2003. ISBN 978-0-12155089-9
- Clayton, Julie. (Ed.). 50 Years of DNA, Palgrave MacMillan Press, 2003. ISBN 978-1-40-391479-8
- Judson, Horace Freeland. The Eighth Day of Creation: Makers of the Revolution in Biology, Cold Spring Harbor Laboratory Press, 1996. ISBN 978-0-87-969478-4
- Olby, Robert. The Path to The Double Helix: Discovery of DNA, first published in October 1974 by MacMillan, with foreword by Francis Crick; ISBN 978-0-48-668117-7; the definitive DNA textbook, revised in 1994, with a 9 page postscript.
- Ridley, Matt. Francis Crick: Discoverer of the Genetic Code (Eminent Lives) HarperCollins Publishers; 192 pp, ISBN 978-0-06-082333-7 2006
- Rose, Steven. The Chemistry of Life, Penguin, ISBN 978-0-14-027273-4.
- Watson, James D. and Francis H.C. Crick. A structure for Deoxyribose Nucleic Acid (PDF). Nature 171, 737–738, 25 April 1953.
- Watson, James D. DNA: The Secret of Life ISBN 978-0-375-41546-3.
- Watson, James D. The Double Helix: A Personal Account of the Discovery of the Structure of DNA (Norton Critical Editions). ISBN 978-0-393-95075-5
- Watson, James D. "Avoid boring people and other lessons from a life in science" (2007) New York: Random House. ISBN 978-0-375-41284-4
External links
- DNA at Curlie
- DNA binding site prediction on protein
- DNA coiling to form chromosomes
- DNA from the Beginning Another DNA Learning Center site on DNA, genes, and heredity from Mendel to the human genome project.
- DNA Lab, demonstrates how to extract DNA from wheat using readily available equipment and supplies.
- DNA the Double Helix Game From the official Nobel Prize web site
- DNA under electron microscope
- Dolan DNA Learning Center
- Double Jelix: 50 years of DNA, Nature
- Double Helix 1953–2003 National Centre for Biotechnology Education
- Francis Crick and James Watson talking on the BBC in 1962, 1972, and 1974
- Genetic Education Modules for Teachers — DNA from the Beginning Study Guide
- Guide to DNA cloning
- Olby, R. (2003) "Quiet debut for the double helix" Nature 421 (January 23): 402–405.
- PDB Molecule of the Month Bci2
- Rosalind Franklin's contributions to the study of DNA
- The Register of Francis Crick Personal Papers 1938 - 2007 at Mandeville Special Collections Library, Geisel Library, University of California, San Diego
- The Secret Life of DNA - DNA Music compositions
- U.S. National DNA Day — watch videos and participate in real-time chat with top scientists
Category:GeneticsCategory:Helices
PM Entry on WikiP
Type of site | Internet encyclopedia project |
|---|---|
| Available in | English |
| Owner | Virginia Tech |
| Created by | Nathan Egge, Aaron Krowne |
| URL | http://planetmath.org/ |
| Commercial | No |
| Registration | Available |
| Current status | active |
PlanetMath is a free, collaborative, online mathematics encyclopedia. The emphasis is on peer review, rigour, openness, pedagogy, real-time content, interlinked content, and community.[1] Intended to be comprehensive, the project is located at the Digital Library Research Lab at Virginia Tech.[2]
PlanetMath was started when the popular free online mathematics encyclopedia MathWorld was taken offline for 12 months by a court injunction as a result of the CRC Press lawsuit against the Wolfram Research company and its employee (and MathWorld's author) Eric Weisstein.[1]
PlanetMath.org website contents
PlanetMath content is licensed under the ‘copyleft’ GNU Free Documentation License.[3] An author who starts a new article becomes the owner of that article; he or she may then choose to grant editing rights to other individuals or groups. All content is written in LaTeX, a typesetting system popular among mathematicians because of its support of the technical needs of mathematical typesetting and its high-quality output. The user can explicitly create links to other articles, and the system also automatically turns certain words into links to the defining articles. The topic area of every article is classified by the Mathematics Subject Classification (MSC) of the American Mathematical Society (AMS).
Users may attach addenda, errata and discussions to articles. A system for private messaging among users is also in place.
As of April 2009, the encyclopedia hosted about 8470 entries and over 14000 concepts (a concept may be e.g. a specific notion defined within a more general entry). About 600 Wikipedia pages incorporate text from PlanetMath and/or PlanetPhysics.org articles [1]
The software running PlanetMath is written in Perl and runs on Linux and the web server Apache. It is known as Noösphere and has been released under the free BSD License.[1]
A PM book of 2,300 pages in PDF format is now available for the PM Free Encyclopedia contents up to 2006 as a free download PDF file of 18Mb [4]. Additional, applied mathematics content[5] andover 2 Gb of bibliographic materials, as well as Encyclopedia entries (currently at >40Mb) related to mathematics applications and Theoretical Physics can be also freely downloaded from the related, but independent, Noosphere-based PlanetPhysics. org website[6][7].
See also
- List of online encyclopedias
- PM Free Encyclopedia v.1.0 Book volumes 1 and 2. 2005., 2305 pages, J. Cornelli and A. Krowne, Eds., GNU License. [8]
- Overview of the current contents of PlanetMath
- Wikipedia:WikiProject Mathematics
- Wikipedia:WikiProject Mathematics/PlanetMath Exchange
- PlanetPhysics.org
- Wikipedia:WikiProject Physics
- Overview of the Contents of PlanetPhysics
- IHES: The fusion of theoretical physics with mathematics at IHES org
- Table of fields of mathematics covered by the PlanetMath.org exchange with Wikipedia
- Higher-dimensional algebra
- Category theory
- Categories, Logic and Foundations of Physics
- Algebraic Topology Foundations of Quantum Field Theory
- Applied mathematics: mathematical physics and physical mathematics
- Stanford Encyclopedia of Philosophy: "Category Theory".
- List of category theory topics
- Important publications in category theory
- Glossary of category theory
- Domain theory
- Enriched category theory
- Higher category theory
- Higher Dimensional Algebra
- Timeline of category theory and related mathematics
- Mathematical biology and Category theory in Biology
- Converting between PlanetMath & PlanetPhysics TeX and Wikipedia mathematical formats.
- Computer graphics in mathematics and physics
- Wikimedia Commons:Pictures and images
- Wikimedia Commons
- MathWorld
References
External links
- PlanetMath
- PlanetPhysics.org
- Categories, Logic and Foundations of Physics
- arXiv
- Archives for Mathematics: arXiv
- Article on PlanetMath in the science magazine of the AAAS
- eprintweb archive
- CogPrints
- Preprint archives at CERN, the EU High Energy Physics International Faciltiy server
- Christoph Lange, "Towards Scientific Collaboration in a Semantic Wiki"
- Aaron E. Klemm, "Motivation and value of free resources: Wikipedia and Planetmath show the way"
- Robert Milson, Aaron Krowne, Adapting CBPP platforms for instructional use
- Mathematics Websites listed on Wikipedia
- Alex M. Andrew, "Archives, mathematics encyclopaedia, dancing robots, ASC", Kybernetes 2008 v. 37: 9/10 pp. 1466 - 1468.
- MathWorld
DNA Molecular Models
Molecular models of DNA structures are representations of the molecular geometry and topology of Deoxyribonucleic acid (DNA) molecules using one of several means, such as: closely packed spheres (CPK models) made of plastic, metal wires for 'skeletal models', graphic computations and animations by computers, artistic rendering, and so on, with the aim of simplifying and presenting the essential, physical and chemical, properties of DNA molecular structures either in vivo or in vitro. Computer molecular models also allow animations and molecular dynamicssimulations that are very important for understanding how DNA functions in vivo. Thus, an old standing dynamic problem is how DNA "self-replication" takes place in living cells that should involve transient uncoiling of supercoiled DNA fibers. Altough DNA consists of relatively rigid, very large elongated biopolymer molecules called "fibers" or chains (that are made of repeating nucleotide units of four basic types, attached to deoxyribose and phospate groups), its molecular stucture in vivo undergoes dynamic configuration changes that involve dynamically attached water molecules and ions. Supercoiling, packing with histones in chromosome structures, and other such supramolecular aspects also involve in vivo DNA topology which is even more complex than DNA molecular geometry, thus turning molecular modeling of DNA into an especially challenging problem for both molecular biologists and biotechnologists. Like other large molecules and biopolymers, DNA often exists in multiple stable geometries (that is, it exhibits conformational isomerism) and configurational, quantum states which are close to each other in energy on the potential energy surface of the DNA molecule. Such geometries can also be computed, at least in principle, by employing ab initio quantum chemistry methods that have high accuracy for small molecules. Such quantum geometries define an important class of ab initio molecular models of DNA whose exploration has barely started.

In an interesting twist of roles, the DNA molecule itself was proposed to be utilized for quantum computing. Both DNA nanostructures as well as DNA 'computing' biochips have been built (see biochip image at right).
The more advanced, computer-based molecular models of DNA involve molecular dynamics simulations as well as quantum mechanical computations of vibro-rotations, delocalized molecular orbitals (MOs), electric dipole moments, hydrogen-bonding, and so on.

Importance
From the very early stages of structural studies of DNA by X-ray diffraction and biochemical means, molecular models such as the Watson-Crick double-helix model were succesfully employed to solve the 'puzzle' of DNA structure, and also find how the latter relates to its key functions in living cells. The first high quality X-ray diffraction patternsof A-DNA were reported by Rosalind Franklin and Raymond Gosling in 1953[1]. The first calculations of the Fourier transform of an atomic helix were reported one year earlier by Cochran, Crick and Vand [2], and were followed in 1953 by the computation of the Fourier transform of a coiled-coil by Crick[3].The first reports of a double-helix molecular model of B-DNA structure were made by Watson and Crick in 1953[4][5]. Last-but-not-least, Maurice F. Wilkins,A. Stokes and H.R. Wilson, reported the first X-ray patternsof in vivo B-DNA in partially oriented salmon sperm heads [6].The development of the first correct double-helix molecular model of DNA by Crick and Watson may not have been possible without the biochemical evidence for the nucleotide base-pairing ([A---T]; [C---G]), or Chargaff's rules[7][8][9][10][11][12].
Examples of DNA molecular models
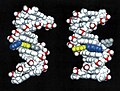
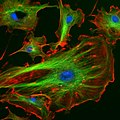
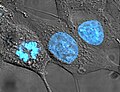
Animated molecular models allow one to visually explore the three-dimensional (3D) structure of DNA. The first DNA model is a space-filling, or CPK, model of the DNA double-helix whereas the third is an animated wire, or skeletal type, molecular model of DNA. The last two DNA molecular models in this series depict quadruplex DNA that may be involved in certain cancers[13][14]. The last figure on this panel is a molecular model of hydrogen bonds between water molecules in ice that are similar to those found in DNA.
- Spacefilling models or CPK models - a molecule is represented by overlapping spheres representing the atoms:
Images for DNA Structure Determination from X-Ray Patterns
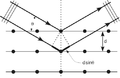
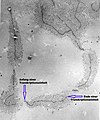
The following images illustrate both the principles and the main steps involved in generating structural information from X-ray diffraction studies of oriented DNA fibers with the help of molecular models of DNA that are combined with crystallographic and mathematical analysis of the X-ray patterns. From left to right the gallery of images shows:
- First row:
- 1. Constructive X-ray interference, or diffraction, following Bragg's Law of X-ray "reflection by the crystal planes";
- 2. A comparison of A-DNA (crystalline) and highly hydrated B-DNA (paracrystalline) X-ray diffraction, and respectively, X-ray scattering patterns (courtesy of Dr. Herbert R. Wilson, FRS- see refs. list);
- 3. Purified DNA precipitated in a water jug;
- 4. The major steps involved in DNA structure determination by X-ray crystallography showing the important role played by molecular models of DNA structure in this iterative, structure--determination process;
- Second row:
- 5. Photo of a modern X-ray diffractometer employed for recording X-ray patterns of DNA with major components: X-ray source, goniometer, sample holder, X-ray detector and/or plate holder;
- 6. Illustrated animation of an X-ray goniometer;
- 7. X-ray detector at the SLAC synchrotron facility;
- 8. Neutron scattering facility at ISIS in UK;
- Third and fourth rows: Molecular models of DNA structure at various scales; figure #11 is an actual electron micrograph of a DNA fiber bundle, presumably of a single bacterial chromosome loop.
Paracrystalline lattice models of B-DNA structures
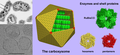
A paracrystalline lattice, or paracrystal,is a molecular or atomic lattice with significant amounts (e.g., larger than a few percent) of partial disordering of molecular arranegements. Limiting cases of the paracrystal model are nanostructures, such as glasses, liquids, etc., that may possess only local ordering and no global order.Liquid crystals also have paracrystalline rather than crystalline structures.
thumb|500px|center|DNA Helix controversy in 1952
Highly hydrated B-DNA occurs naturally in living cells in such a paracrystalline state, which is a dynamic one in spite of the relatively rigid DNA double-helix stabilized by parallel hydrogen bonds between the nucleotide base-pairs in the two complementary, helical DNA chains (see figures). For simplicity most DNA molecular models ommit both water and ions dynamically bound to B-DNA, and are thus less useful for understanding the dynamic behaviors of B-DNA in vivo. The physical and mathematical analysis of X-ray[15][16] and spectroscopic data for paracrystalline B-DNA is therefore much more complicated than that of crystalline, A-DNA X-ray diffraction patterns. The paracrystal model is also important for DNA technological applications such as DNA nanotechnology. Novel techniques that combine X-ray diffraction of DNA with X-ray microscopy in hydrated living cells are now also being developed (see, for example, "Application of X-ray microscopy in the analysis of living hydrated cells").
Genomic and Biotechnology Applications of DNA molecular modeling
The following gallery of images illustrates various uses of DNA molecular modeling in Genomics and Biotechnology research applications from DNA repair to PCR and DNA nanostructures; each slide contains its own explanation and/or details. The first slide presents an overview of DNA applications, including DNA molecular models, with emphasis on Genomics and Biotechnology.
Gallery: DNA Molecular modeling applications
Databases for DNA molecular models and sequences
X-ray diffraction
- NDB ID: UD0017 Database
- X-ray Atlas -database
- PDB files of coordinates for nucleic acid structures from X-ray diffraction by NA (incl. DNA) crystals
- Structure factors dowloadable files in CIF format
Neutron scattering
- ISIS neutron source
- ISIS pulsed neutron source:A world centre for science with neutrons & muons at Harwell, near Oxford, UK.
X-ray microscopy
Electron microscopy
Atomic Force Microscopy (AFM)
Two-dimensional DNA junction arrays have been visualized by Atomic Force Microscopy (AFM)[17]. Other imaging resources for AFM/Scanning probe microscopy(SPM) can be freely accessed at:
Gallery of AFM Images
Mass spectrometry--Maldi informatics
Spectroscopy
- Vibrational circular dichroism (VCD)
- FT-NMR[18][19]
- NMR microscopy[20]
- Microwave spectroscopy
- FT-IR
- FT-NIR[21][22][23]
- Spectral, Hyperspectral, and Chemical imaging)[24][25][26][27][28][29][30].
- Raman spectroscopy/microscopy[31] and CARS[32].
- Fluorescence correlation spectroscopy[33][34][35][36][37][38][39][40], Fluorescence cross-correlation spectroscopy and FRET[41][42][43].
- Confocal microscopy[44]
Gallery: CARS (Raman spectroscopy), Fluorescence confocal microscopy, and Hyperspectral imaging
Genomic and structural databases
- CBS Genome Atlas Database — contains examples of base skews.[45]
- The Z curve database of genomes — a 3-dimensional visualization and analysis tool of genomes[46].
- DNA and other nucleic acids' molecular models: Coordinate files of nucleic acids molecular structure models in PDB and CIF formats
Notes
References
- Applications of Novel Techniques to Health Foods, Medical and Agricultural Biotechnology.(June 2004) I. C. Baianu, P. R. Lozano, V. I. Prisecaru and H. C. Lin., q-bio/0406047.
- F. Bessel, Untersuchung des Theils der planetarischen Störungen, Berlin Abhandlungen (1824), article 14.
- Sir Lawrence Bragg, FRS. The Crystalline State, A General survey. London: G. Bells and Sons, Ltd., vols. 1 and 2., 1966., 2024 pages.
- Cantor, C. R. and Schimmel, P.R. Biophysical Chemistry, Parts I and II., San Franscisco: W.H. Freeman and Co. 1980. 1,800 pages.
- Eigen, M., and Rigler, R. (1994). Sorting single molecules: Applications to diagnostics and evolutionary biotechnology, Proc. Natl. Acad. Sci. USA 91:5740.
- Raghavachari, R., Editor. 2001. Near-Infrared Applications in Biotechnology, Marcel-Dekker, New York, NY.
- Rigler R. and Widengren J. (1990). Ultrasensitive detection of single molecules by fluorescence correlation spectroscopy, BioScience (Ed. Klinge & Owman) p.180.
- Single Cancer Cell Detection by Near Infrared Microspectroscopy, Infrared Chemical Imaging and Fluorescence Microspectroscopy.2004. I. C. Baianu, D. Costescu, N. E. Hofmann, S. S. Korban and et al., q-bio/0407006 (July 2004).
- Voet, D. and J.G. Voet. Biochemistry, 2nd Edn., New York, Toronto, Singapore: John Wiley & Sons, Inc., 1995, ISBN: 0-471-58651-X., 1361 pages.
- Watson, G. N. A Treatise on the Theory of Bessel Functions., (1995) Cambridge University Press. ISBN 0-521-48391-3.
- Watson, James D. and Francis H.C. Crick. A structure for Deoxyribose Nucleic Acid (PDF). Nature 171, 737–738, 25 April 1953.
- Watson, James D. Molecular Biology of the Gene. New York and Amsterdam: W.A. Benjamin, Inc. 1965., 494 pages.
- Wentworth, W.E. Physical Chemistry. A short course., Malden (Mass.): Blackwell Science, Inc. 2000.
- Herbert R. Wilson, FRS. Diffraction of X-rays by proteins, Nucleic Acids and Viruses., London: Edward Arnold (Publishers) Ltd. 1966.
- Kurt Wuthrich. NMR of Proteins and Nucleic Acids., New York, Brisbane,Chicester, Toronto, Singapore: J. Wiley & Sons. 1986., 292 pages.
- Robinson, Bruche H. (1987). "The Design of a Biochip: A Self-Assembling Molecular-Scale Memory Device". Protein Engineering. 1 (4): 295–300. ISSN 0269-2139.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) Link - Rothemund, Paul W. K. (2004). "Design and Characterization of Programmable DNA Nanotubes". Journal of the American Chemical Society. 126 (50): 16344–16352. doi:10.1021/ja044319l. ISSN 0002-7863.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - Keren, K. (2003). "DNA-Templated Carbon Nanotube Field-Effect Transistor". Science. 302 (6549): 1380–1382. doi:10.1126/science.1091022. ISSN 1095-9203.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - Zheng, Jiwen (2006). "2D Nanoparticle Arrays Show the Organizational Power of Robust DNA Motifs". Nano Letters. 6: 1502–1504. doi:10.1021/nl060994c. ISSN 1530-6984.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - Cohen, Justin D. (2007). "Addressing Single Molecules on DNA Nanostructures". Angewandte Chemie. 46 (42): 7956–7959. doi:10.1002/anie.200702767. ISSN 0570-0833.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - Mao, Chengde (1999). "Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy". Journal of the American Chemical Society. 121 (23): 5437–5443. doi:10.1021/ja9900398. ISSN 0002-7863.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - Constantinou, Pamela E. (2006). "Double cohesion in structural DNA nanotechnology". Organic and Biomolecular Chemistry. 4: 3414–3419. doi:10.1039/b605212f.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)
See also
- DNA
- Molecular graphics
- DNA structure
- X-ray scattering
- Neutron scattering
- Crystallography
- Crystal lattices
- Paracrystalline lattices/Paracrystals
- 2D-FT NMRI and Spectroscopy
- NMR Spectroscopy
- Microwave spectroscopy
- Two-dimensional IR spectroscopy
- Spectral imaging
- Hyperspectral imaging
- Chemical imaging
- NMR microscopy
- VCD or Vibrational circular dichroism
- FRET and FCS- Fluorescence correlation spectroscopy
- Fluorescence cross-correlation spectroscopy (FCCS)
- Molecular structure
- Molecular geometry
- Molecular topology
- DNA topology
- Nanostructure
- DNA nanotechnology
- Imaging
- Atomic force microscopy
- X-ray microscopy
- Liquid crystal
- Glasses
- QMC@Home
- Sir Lawrence Bragg, FRS
- Sir John Randall
- James Watson
- Francis Crick
- Maurice Wilkins
- Herbert Wilson, FRS
- Alex Stokes
External links
- DNA the Double Helix Game From the official Nobel Prize web site
- MDDNA: Structural Bioinformatics of DNA
- Double Helix 1953–2003 National Centre for Biotechnology Education
- DNA under electron microscope
- Ascalaph DNA — Commercial software for DNA modeling
- DNAlive: a web interface to compute DNA physical properties. Also allows cross-linking of the results with the UCSC Genome browser and DNA dynamics.
- DiProDB: Dinucleotide Property Database. The database is designed to collect and analyse thermodynamic, structural and other dinucleotide properties.
- Further details of mathematical and molecular analysis of DNA structure based on X-ray data
- Bessel functions corresponding to Fourier transforms of atomic or molecular helices.
- Application of X-ray microscopy in analysis of living hydrated cells
- Characterization in nanotechnology some pdfs
- overview of STM/AFM/SNOM principles with educative videos
- SPM Image Gallery - AFM STM SEM MFM NSOM and More
- How SPM Works
- U.S. National DNA Day — watch videos and participate in real-time discusssions with scientists.
- The Secret Life of DNA - DNA Music compositions
Category:Molecular geometryCategory:HelicesCategory:DiffractionCategory:Molecular dynamicsCategory:ScatteringCategory:Quantum chemistryCategory:Computational modelsCategory:Molecular biologyCategory:Molecular geneticsCategory:GenomicsCategory:GeneticsCategory:Crystallography Category:Spectroscopy *Category:PolymersCategory:BiomoleculesCategory:NanotechnologyCategory:Biotechnology productsCategory:DatabasesCategory:Microscopic imagesCategory:Scanning probe microscopyCategory:FluorescenceCategory:Database management systemsCategory:Classes of computersCategory:Information theoryCategory:Nobel laureates in Physiology or MedicineCategory:Molecular biologistsCategory:Biophysicists
Projects
- WP-Math
- WPBiography
Complex Systems Biology and Complexity Science
The Complexity science field is the new science field about complexity, which treats complex systems as a field of science. This scientific field studies the common properties of systems that are considered fundamentally complex. Such systems may exist in nature, society, science and other many fields. It is also called complex systems theory, complexity science, study of complex systems, sciences of complexity, non-equilibrium physics, and historical physics.The key problems of such systems are difficulties with their formal modeling and simulation. From such perspective, in different research contexts complex systems are defined on the base of their different attributes.At present, the consensus related to one universal definition of complex system does not exist yet.
Overview

In these endeavors, scientists often seek simple non-linear coupling rules which lead to complex phenomena (rather than describe - see above), but this need not be the case. Human societies (and probably human brains) are complex systems in which neither the components nor the couplings are simple. Nevertheless, they exhibit many of the hallmarks of complex systems. It is worth remarking that non-linearity is not a necessary feature of complex systems modeling: macro-analyses that concern unstable equilibrium and evolution processes of certain biological/social/economic systems can usefully be carried out also by sets of linear equations, which do nevertheless entail reciprocal dependence between variable parameters.
Traditionally, engineering has striven to keep its systems linear, because that makes them simpler to build and to predict. However, many physical systems (for example lasers) are inherently "complex systems" in terms of the definition above, and engineering practice must now include elements of complex systems research.
Information theory applies well to the complex adaptive systems, CAS, through the concepts of object oriented design, as well as through formalized concepts of organization and disorder that can be associated with any systems evolution process.
History
Complex Systems is a new approach to science that studies how relationships between parts give rise to the collective behaviors of a system and how the system interacts and forms relationships with its environment.
The earliest precursor to modern complex systems theory can be found in the classical political economy of the Scottish Enlightenment, later developed by the Austrian school of economics, which says that order in market systems is spontaneous (or emergent) in that it is the result of human action, but not the execution of any human design.[2][3]
Upon this the Austrian school developed from the 19th to the early 20th century the economic calculation problem, along with the concept of dispersed knowledge, which were to fuel debates against the then-dominant Keynesian economics. This debate would notably lead economists, politicians and other parties to explore the question of computational complexity.
A pioneer in the field, and inspired by Karl Popper's and Warren Weaver's works, Nobel prize economist and philosopher Friedrich Hayek dedicated much of his work, from early to the late 20th century, to the study of complex phenomena,[4] not constraining his work to human economies but to other fields such as psychology,[5] biology and cybernetics.
Further Steven Strogatz from Sync stated that "every decade or so, a grandiose theory comes along, bearing similar aspirations and often brandishing an ominous-sounding C-name. In the 1960s it was cybernetics. In the '70s it was catastrophe theory. Then came chaos theory in the '80s and complexity theory in the '90s."
Topics in the complex systems study
Complexity and modeling

Complexity and chaos theory
Complexity theory is rooted in Chaos theory, which in turn has its origins more than a century ago in the work of the French mathematician Henri Poincaré. Chaos is sometimes viewed as extremely complicated information, rather than as an absence of order.[8] The point is that chaos remains deterministic. With perfect knowledge of the initial conditions and of the context of an action, the course of this action can be predicted in chaos theory. As argued by Ilya Prigogine,[9] Complexity is non-deterministic, and gives no way whatsoever to predict the future. The emergence of complexity theory shows a domain between deterministic order and randomness which is complex.[10] This is referred as the 'edge of chaos'.[11]
When one analyses complex systems, sensitivity to initial conditions, for example, is not an issue as important as within the chaos theory in which it prevails. As stated by Colander,[12] the study of complexity is the opposite of the study of chaos. Complexity is about how a huge number of extremely complicated and dynamic set of relationships can generate some simple behavioural patterns, whereas chaotic behaviour, in the sense of deterministic chaos, is the result of a relatively small number of non-linear interactions.[10]
Therefore, the main difference between Chaotic systems and complex systems is their history.[13] Chaotic systems don’t rely on their history as complex ones do. Chaotic behaviour pushes a system in equilibrium into chaotic order, which means, in other words, out of what we traditionally define as 'order'. On the other hand, complex systems evolve far from equilibrium at the edge of chaos. They evolve at a critical state built up by a history of irreversible and unexpected events. In a sense chaotic systems can be regarded as a subset of complex systems distinguished precisely by this absence of historical dependence. Many real complex systems are, in practice and over long but finite time periods, robust. However, they do possess the potential for radical qualitative change of kind whilst retaining systemic integrity. Metamorphosis serves as perhaps more than a metaphor for such transformations.
Research centers, conferences, and journals
Institutes and research centers
- New England Complex Systems Institute
- Santa Fe Institute
- Center for Social Dynamics & Complexity (CSDC) at Arizona State University
- Southampton Institute for Complex Systems Simulation
- Center for the Study of Complex Systems at the University of Michigan
- Center for Complex Systems and Brain Sciences at Florida Atlantic University
Journals
- Complex Systems journal
- Interdisciplinary Description of Complex Systems journal
See also
References
Further reading
- L.A.N. Amaral and J.M. Ottino, Complex networks — augmenting the framework for the study of complex system, 2004.
- Murray Gell-Mann, Let's Call It Plectics, 1995/96.
- Nigel Goldenfeld and Leo P. Kadanoff, Simple Lessons from Complexity, 1999
- A. Gogolin, A. Nersesyan and A. Tsvelik, Theory of strongly correlated systems , Cambridge University Press, 1999.
- Kelly, K. (1995). Out of Control, Perseus Books Group.
- Graeme Donald Snooks, "A general theory of complex living systems: Exploring the demand side of dynamics", Complexity, vol. 13, no. 6, July/August 2008.
- Sorin Solomon and Eran Shir, Complexity; a science at 30, 2003.
External links
Page categories
Reference list
testnu mediawiki
test page with dna image