The Gamma variant (P.1)[a] was[6][7] one of the variants of SARS-CoV-2, the virus that causes COVID-19.[8] This variant of SARS-CoV-2 has been named lineage P.1 and has 17 amino acid substitutions, ten of which in its spike protein, including these three designated to be of particular concern: N501Y, E484K and K417T.[4][9] It was first detected by the National Institute of Infectious Diseases (NIID) of Japan, on 6 January 2021 in four people who had arrived in Tokyo having visited Amazonas, Brazil, four days earlier.[4][10] It was subsequently declared to be in circulation in Brazil.[4] Under the simplified naming scheme proposed by the World Health Organization, P.1 was labeled Gamma variant, and was considered a variant of concern until March 2022, when it was largely displaced by the delta and omicron variants.[11]
| Gamma | |
 | |
| General details | |
|---|---|
| WHO Designation | Gamma |
| Other Names | 20J/501Y.V3, Variant of Concern 202101/02 (VOC-202101/02),[1] Brazilian variant or Brazil variant'[2][3][4] |
| Lineage | P.1 |
| First detected | Tokyo, Japan |
| Date reported | 6 January 2021 |
| Status | Variant of concern |
| Cases map | |
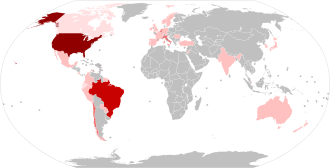 Legend: 1,000+ confirmed sequences 500–999 confirmed sequences 100–499 confirmed sequences 2–99 confirmed sequences 1 confirmed sequence None or no data available | |
| Major variants | |
Gamma caused widespread infection in early 2021 in the city of Manaus, the capital of Amazonas, although the city had already experienced widespread infection in May 2020,[12] with a study[13] indicating high seroprevalence of antibodies for SARS-CoV-2.[14] A research article published in Science Journal indicate that P.1 infected people have a greater chance of transmissibility and death than B.1.1.28 infected ones.[15]
The Gamma variant comprises the two distinct subvariants 28-AM-1 and 28-AM-2, which both carry the K417T, E484K, N501Y mutations, and which both developed independently of each other within the same Brazilian Amazonas region.[16]
Gamma is notably different from the Zeta variant (lineage P.2) which also circulated strongly in Brazil. In particular, Zeta only carries the E484K mutation and has neither of the other two mutations of concern, N501Y and K417T.[16][9]
Classification
Initial reports claimed that both P.1 and P.2 were two separate and different descendants of the Brazilian lineage B.1.1.248.[17][18] However, B.1.1.248 later lost its status as a distinct lineage and was reclassified to B.1.1.28.[19] P.1 has also been called B.1.1.28.1,[20] while P.2 has been B.1.1.28.2 or VUI-202101/01.[21] Since only three sublevels are permitted in the PANGO Lineage system of nomenclature, hence the designation of B.1.1.28.1 to P.1 and B.1.1.28.2 to P.2.[4][22]
Following its detection, genome data for four samples of the new variant were shared to GISAID having been assigned the ID range: EPI_ISL_792680 to EPI_ISL_792683.[23]
Mutations
| Gene | Amino acid |
|---|---|
| ORF1ab | synT733C |
| synC2749T | |
| S1188L | |
| K1795Q | |
| del11288-11296 (3675-3677 SGF) | |
| synC12778T | |
| synC13860T | |
| E5665D | |
| Spike | L18F |
| T20N | |
| P26S | |
| D138Y | |
| R190S | |
| K417T | |
| E484K | |
| N501Y | |
| H 655Y | |
| T1027I | |
| ORF8 | E92K |
| ins28269-28273 | |
| N | P80R |
| Source: Faria et al. (2021), Figure 1.B | |
Variants of SARS-CoV-2
As well as having eight mutations (four of these synonymous genetic mutations) in its open reading frames (ORF1a and ORF1b) – one of which is a set of deletions – Gamma has 10 defining mutations in its spike protein, including N501Y and E484K. It also has two mutations – one an insertion – in its ORF8 gene and one in its N gene.[4][24]
Descendant and sublineages
Coronavirus lineage B.1.1.28 has originated four known lineages classified as variant of interest (VOI) or variant of concern (VOC): lineages P.1, P.2, P.3 and P.4.
Lineage P.2 (B.1.1.28.2, Zeta variant), first detected in October 2020 in the state of Rio de Janeiro, Brazil, only shares one mutation of concern with P.1, which is the E484K.[26] The other P.2 mutations are without concern and rarely found for other variants. The five P.2-specific mutations are: E484K in S-gene, A119S in N-gene, 5’UTR C100U, plus L3468V and synC11824U in ORF1ab-gene. Other mutations commonly found in P.2 are: 3’UTR C29754U, F120F (synC28253U) in ORF8, M234I in the N-gene, plus L3930F and synA12964G in ORF1ab.[27]
Lineage P.3 (Theta variant) was first identified in the Philippines on 18 February 2021 when two mutations of concern were detected in Central Visayas.[28]
The remaining B.1.1.28 derivative virus is lineage P.4. Although researchers have not identified its precise origin, it was first sequenced in Itirapina, Brazil, and was already circulating in various municipalities in the state of São Paulo of the same country. It carries a mutation of concern in the spike protein called L452R which is also present in lineage B.1.617 (Delta and Kappa variants) detected in India, Epsilon variant (lineages B.1.427 and B.1.429) from California, United States.[29][30] The branch of this lineage is P.4.1 (VUI-NP13L)—suspected to have arisen in Goiás, Brazil, around June–July 2020— also rapidly spread to the southeast of the country, where for example Taquara had its first genome sequence, and to the northeast of the nation. It was detected internationally, with reported cases in Japan, Netherlands and England. The P.4.1 has V1176F and D614G mutations in spike protein.[31]
Prevention
Statistics
| Country | Confirmed cases (PANGO) | Confirmed cases (GISAID)[32] as of 3 November 2023 | First detection | References |
|---|---|---|---|---|
 United States United States | 15,786 | 23,373 | 25 January 2021 | [33][34][35][36] |
 Canada Canada | 19,643 | 8,070 | 7 February 2021 | [36][37][38][39] |
 Brazil Brazil | 10,556 | 16,200 | 14 January 2021 | [40][4][36] |
 Chile Chile | 975 | 2,522 | 24 March 2021 | [41][36] |
 Mexico Mexico | 814 | 2,278 | 28 January 2021 | [36] |
 Italy Italy | 838 | 2,181 | 25 January 2021 | [42][43][44][36] |
 Belgium Belgium | 1,333 | 1,974 | 16 February 2021 | [36] |
 Spain Spain | 635 | 957 | 16 February 2021 | [36] |
 Luxembourg Luxembourg | 43 | 928 | 30 March 2021 | [36] |
 Germany Germany | 276 | 806 | 22 January 2021 | [45][36] |
 France France | 219 | 577 | 4 February 2021 | [36][46] |
 Netherlands Netherlands | 499 | 566 | 29 January 2021 | [47][36] |
 Colombia Colombia | 165 | 346 | 30 January 2021 | [48][49][36] |
 Argentina Argentina | 230 | 329 | 8 February 2021 | [50][36] |
 French Guiana French Guiana | 225 | 318 | 16 February 2021 | [36] |
 Trinidad and Tobago Trinidad and Tobago | 14 | 255 | 23 April 2021 | [51][52][53] |
 United Kingdom United Kingdom | 155 | 224 | 28 February 2021 | [36][54][55][56][1] |
 Portugal Portugal | 148 | 190 | 11 February 2021 | [57][58][59][36] |
 Switzerland Switzerland | 153 | 187 | 16 February 2021 | [36] |
 Uruguay Uruguay | 112 | 173 | 22 March 2021 | [60] |
 Turkey Turkey | 166 | 88 | 3 February 2021 | [61][62][36][63] |
 Ecuador Ecuador | 27 | 145 | 14 April 2021 | [36] |
 Sweden Sweden | 68 | 141 | 20 February 2021 | [64][36] |
 Aruba Aruba | 96 | 121 | 29 March 2021 | [36] |
 Japan Japan | 96 | 118 | 6 January 2021 | [65][66][36] |
 Suriname Suriname | 106 | 117 | 11 April 2021 | [36] |
 Peru Peru | 103 | 42 | 4 February 2021 | [67][68][69][70][71] |
 Costa Rica Costa Rica | 32 | 69 | 11 April 2021 | [36] |
 Denmark Denmark | 40 | 63 | 3 March 2021 | [72][73][36] |
 Paraguay Paraguay | 53 | 54 | 25 March 2021 | [74][36] |
 Haiti Haiti | 46 | 47 | 15 May 2021 | [36] |
 Dominican Republic Dominican Republic | 4 | 38 | 11 March 2021 | [36] |
 Malta Malta | 22 | 32 | 18 April 2021 | [36] |
 Ireland Ireland | 25 | 29 | 19 February 2021 | [75][76][77][78][79][36] |
 Austria Austria | 17 | 28 | 15 April 2021 | [36] |
 Poland Poland | 5 | 24 | 13 April 2021 | [36] |
 Czech Republic Czech Republic | 9 | 20 | 21 April 2021 | [36] |
 Israel Israel | 9 | 19 | 11 April 2021 | [36] |
 Venezuela Venezuela | 17 | 17 | 10 February 2021 | [36] |
 Bolivia Bolivia | 10 | 17 | 19 March 2021 | [36] |
 Curaçao Curaçao | 6 | 14 | 24 April 2021 | [36] |
 Romania Romania | 5 | 12 | 8 March 2021 | [80][36] |
 South Korea South Korea | 3 | 10 | 18 January 2021 | [81] |
 Norway Norway | 4 | 9 | 24 February 2021 | [36] |
 Singapore Singapore | 8 | 8 | 16 April 2021 | [36] |
 Australia Australia | 5 | 8 | 6 March 2021 | [36] |
 New Zealand New Zealand | 7 | 7 | 18 March 2021 | [36] |
 Slovenia Slovenia | 4 | 7 | 21 March 2021 | [36] |
 Finland Finland | 2 | 7 | 18 February 2021 | [82] |
 Lithuania Lithuania | 3 | 6 | 10 May 2021 | [36] |
 Croatia Croatia | 2 | 6 | 18 February 2021 | [83] |
 Jordan Jordan | 5 | 5 | 4 April 2021 | [36] |
 Taiwan Taiwan | 4 | 4 | 20 February 2021 | [36] |
 India India | 2 | 3 | 17 February 2021 | [84] |
 Bosnia and Herzegovina Bosnia and Herzegovina | 1 | 3 | ||
 Guyana Guyana | 1 | 3 | ||
 China China | 2 | 2 | 10 May 2021 | [36] |
 Philippines Philippines | 2 | 2 | 13 March 2021 | [85][86] |
 Bangladesh Bangladesh | 1 | 1 | 3 May 2021 | [36] |
 Cayman Islands Cayman Islands | 1 | 1 | 17 June 2021 | [36] |
 Faroe Islands Faroe Islands | 1 | 1 | 18 January 2021 | [87][36] |
 Thailand Thailand | 1 | 1 | 5 April 2021 | [36] |
 Guyane Guyane | 1 | 1 | 3 May 2021 | [36] |
 Hungary Hungary | 1 | 1 | 22 July 2021 | [88] |
 Sint Maarten Sint Maarten | 1 | 1 | 10 March 2021 | [36] |
 Angola Angola | 1 | 1 | ||
 Bonaire Bonaire | 1 | 1 | ||
 Greece Greece | 1 | 1 | ||
 Guam Guam | 1 | 1 | ||
 Latvia Latvia | 1 | 1 | ||
 Pakistan Pakistan | 1 | 1 | ||
 Puerto Rico Puerto Rico | 1 | 1 | ||
 Russia Russia | 1 | 1 | ||
| World (63 countries) | Total: 52,079 | Total: 63,729 |
History
On 12 January 2021, the Brazil–United Kingdom CADDE Centre confirmed 13 local cases of lineage P.1 in Manaus, Amazonas state, the largest city of the Amazon rain forest.[4] The new lineage was absent in 27 samples collected from March to November 2020 from Manaus, but it was identified for the same city in 42% (n=13/31) of the samples collected 15–23 December 2020, followed by 52.2% (n=35/67) during 15–31 December 2020 and 85.4% (n=41/48) during 1–9 January 2021. Most notably, the P.2 was rapidly outcompeted by P1 going from the second half of December to 1–9 January, where the lineage P.2 share for Manaus decreased from 25.4% to 6.3%.[4][89]
A study of 180 sequenced Brazilian samples collected in the state of Rio de Janeiro during 2020, identified emergence of the novel lineage P.2 of SARS-CoV-2 (originating from B.1.1.28). P.2 was first detected by genome sequencing in October 2020, but it was estimated to have emerged in early July 2020.[27] As of December 2020, although having significantly increased in frequency throughout the state, it was still largely confined to the state capital Rio de Janeiro. In May 2020 the main lineages behind the COVID-19 positives were B.1.1.33 (70%) and B.1.1.28 (20%), whereas by September the main lineages were B.1.1.33 (50%) and B.1.1.28 (40%), with no detected presence of P.2, while during October and November P.2 was the most common lineage with a share close to 50% (according to the Pangolin tool).[90] The study also found the E484K mutation as "widely spread" across all analysed P.2 samples (36 out of 38).[90]
Researchers at the Oswaldo Cruz Foundation published a preprint genomic epidemiology study of 250 collected genomes from different places in Amazonas and found that P.1 infections can produce nearly 10 times more viral load than in other COVID-19-infected persons involving lineages B.1.1.28 and B.1.195. The lineage also showed 2.2 times higher transmissibility with the same ability to infect both adults (18–59 years old) and older persons (60 years old and higher), suggesting P.1 and its sublineages are more successful at infecting younger humans with no gender differential.[91]
The Centre for Arbovirus Discovery, Diagnosis, Genomics and Epidemiology (CADDE) produced another journal article of samples collected in Manaus between November 2020 and January 2021. The study indicated lineage P.1 to be about 2.0 times (50% CrI, 1.7–2.4 times) more transmissible and was shown to be capable of evading about 32% (50% CrI, 21–46%) of inherited immunity from previous coronavirus diseases, leading to the possibility of reinfection. These increased statistics also had the same reflection in fatality, in that P.1 infections can be about 50% (50% CrI, 20–90%) more lethal.[92][93][94] As part of ongoing research, the variant's capacity to neutralise antibodies has been evaluated by scientists in a published preprint work demonstrating that 8 CoronaVac-immunised persons had a poor blood plasma response against lineage P.1. Since the study only had a small number of participants, it was not possible to establish any statistical conclusion as a larger number of vaccinated people would need to be studied.[95] Scientists at MIT, Harvard and Cambridge, and hospitals physicians in Boston, corroborated that people fully vaccinated with Pfizer and Moderna vaccines have significantly decreased neutralisation with P.1—in a preprint work.[96]
Extinction
In March 2022, the World Health Organization listed the Alpha, Beta and Gamma variants as previously circulating citing lack of any detected cases in the prior weeks and months.[11]



